Muhammad Febrian Rachmadi
Few-shot medical image classification with simple shape and texture text descriptors using vision-language models
Aug 08, 2023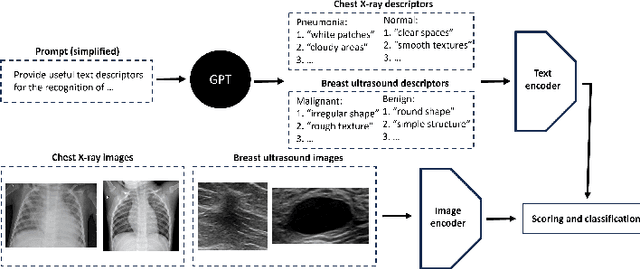
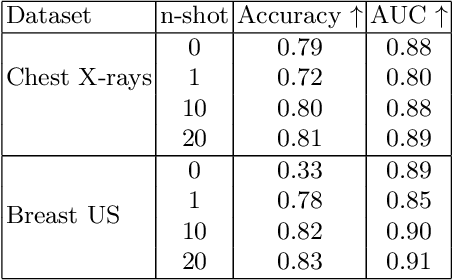
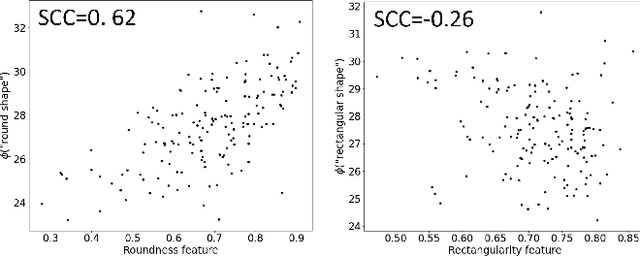

Abstract:In this work, we investigate the usefulness of vision-language models (VLMs) and large language models for binary few-shot classification of medical images. We utilize the GPT-4 model to generate text descriptors that encapsulate the shape and texture characteristics of objects in medical images. Subsequently, these GPT-4 generated descriptors, alongside VLMs pre-trained on natural images, are employed to classify chest X-rays and breast ultrasound images. Our results indicate that few-shot classification of medical images using VLMs and GPT-4 generated descriptors is a viable approach. However, accurate classification requires to exclude certain descriptors from the calculations of the classification scores. Moreover, we assess the ability of VLMs to evaluate shape features in breast mass ultrasound images. We further investigate the degree of variability among the sets of text descriptors produced by GPT-4. Our work provides several important insights about the application of VLMs for medical image analysis.
White Matter Hyperintensities Segmentation Using Probabilistic TransUNet
May 06, 2023Abstract:White Matter Hyperintensities (WMH) are areas of the brain that have higher intensity than other normal brain regions on Magnetic Resonance Imaging (MRI) scans. WMH is often associated with small vessel disease in the brain, making early detection of WMH important. However, there are two common issues in the detection of WMH: high ambiguity and difficulty in detecting small WMH. In this study, we propose a method called Probabilistic TransUNet to address the precision of small object segmentation and the high ambiguity of medical images. To measure model performance, we conducted a k-fold cross validation and cross dataset robustness experiment. Based on the experiments, the addition of a probabilistic model and the use of a transformer-based approach were able to achieve better performance.
Improving Segmentation of Objects with Varying Sizes in Biomedical Images using Instance-wise and Center-of-Instance Segmentation Loss Function
Apr 13, 2023



Abstract:In this paper, we propose a novel two-component loss for biomedical image segmentation tasks called the Instance-wise and Center-of-Instance (ICI) loss, a loss function that addresses the instance imbalance problem commonly encountered when using pixel-wise loss functions such as the Dice loss. The Instance-wise component improves the detection of small instances or ``blobs" in image datasets with both large and small instances. The Center-of-Instance component improves the overall detection accuracy. We compared the ICI loss with two existing losses, the Dice loss and the blob loss, in the task of stroke lesion segmentation using the ATLAS R2.0 challenge dataset from MICCAI 2022. Compared to the other losses, the ICI loss provided a better balanced segmentation, and significantly outperformed the Dice loss with an improvement of $1.7-3.7\%$ and the blob loss by $0.6-5.0\%$ in terms of the Dice similarity coefficient on both validation and test set, suggesting that the ICI loss is a potential solution to the instance imbalance problem.
Hierarchical Vision Transformers for Cardiac Ejection Fraction Estimation
Mar 31, 2023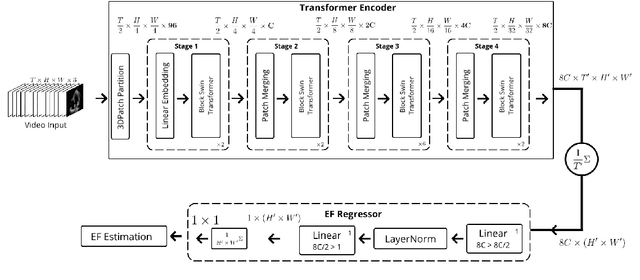
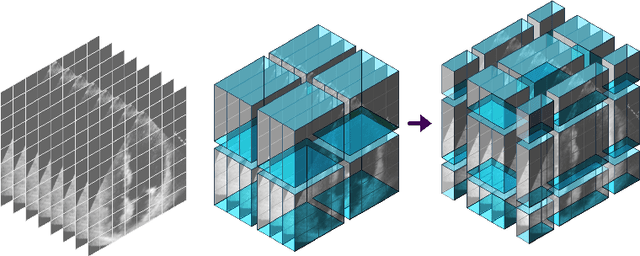
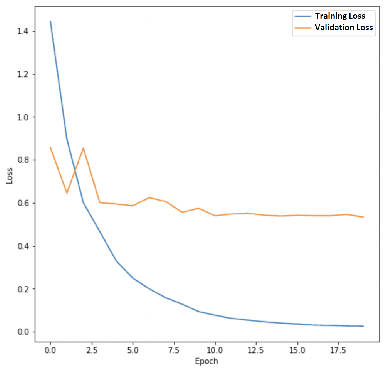
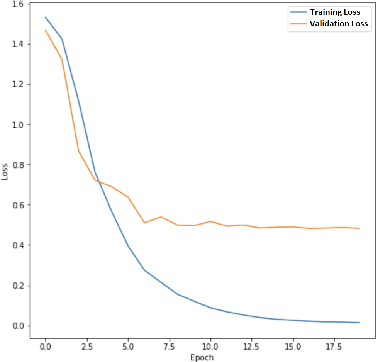
Abstract:The left ventricular of ejection fraction is one of the most important metric of cardiac function. It is used by cardiologist to identify patients who are eligible for lifeprolonging therapies. However, the assessment of ejection fraction suffers from inter-observer variability. To overcome this challenge, we propose a deep learning approach, based on hierarchical vision Transformers, to estimate the ejection fraction from echocardiogram videos. The proposed method can estimate ejection fraction without the need for left ventrice segmentation first, make it more efficient than other methods. We evaluated our method on EchoNet-Dynamic dataset resulting 5.59, 7.59 and 0.59 for MAE, RMSE and R2 respectivelly. This results are better compared to the state-of-the-art method, Ultrasound Video Transformer (UVT). The source code is available on https://github.com/lhfazry/UltraSwin.
An automated pipeline to create an atlas of in situ hybridization gene expression data in the adult marmoset brain
Mar 13, 2023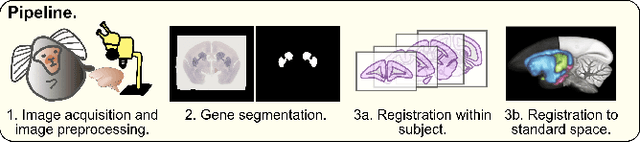
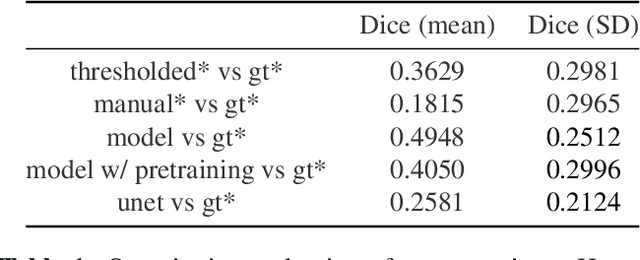
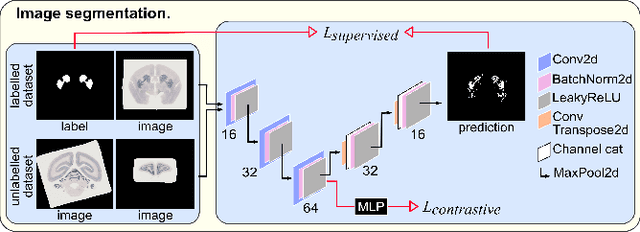

Abstract:We present the first automated pipeline to create an atlas of in situ hybridization gene expression in the adult marmoset brain in the same stereotaxic space. The pipeline consists of segmentation of gene expression from microscopy images and registration of images to a standard space. Automation of this pipeline is necessary to analyze the large volume of data in the genome-wide whole-brain dataset, and to process images that have varying intensity profiles and expression patterns with minimal human bias. To reduce the number of labelled images required for training, we develop a semi-supervised segmentation model. We further develop an iterative algorithm to register images to a standard space, enabling comparative analysis between genes and concurrent visualization with other datasets, thereby facilitating a more holistic understanding of primate brain structure and function.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge