Charissa Poon
Implicit neural representations for joint decomposition and registration of gene expression images in the marmoset brain
Aug 08, 2023Abstract:We propose a novel image registration method based on implicit neural representations that addresses the challenging problem of registering a pair of brain images with similar anatomical structures, but where one image contains additional features or artifacts that are not present in the other image. To demonstrate its effectiveness, we use 2D microscopy $\textit{in situ}$ hybridization gene expression images of the marmoset brain. Accurately quantifying gene expression requires image registration to a brain template, which is difficult due to the diversity of patterns causing variations in visible anatomical brain structures. Our approach uses implicit networks in combination with an image exclusion loss to jointly perform the registration and decompose the image into a support and residual image. The support image aligns well with the template, while the residual image captures individual image characteristics that diverge from the template. In experiments, our method provided excellent results and outperformed other registration techniques.
Improving Segmentation of Objects with Varying Sizes in Biomedical Images using Instance-wise and Center-of-Instance Segmentation Loss Function
Apr 13, 2023



Abstract:In this paper, we propose a novel two-component loss for biomedical image segmentation tasks called the Instance-wise and Center-of-Instance (ICI) loss, a loss function that addresses the instance imbalance problem commonly encountered when using pixel-wise loss functions such as the Dice loss. The Instance-wise component improves the detection of small instances or ``blobs" in image datasets with both large and small instances. The Center-of-Instance component improves the overall detection accuracy. We compared the ICI loss with two existing losses, the Dice loss and the blob loss, in the task of stroke lesion segmentation using the ATLAS R2.0 challenge dataset from MICCAI 2022. Compared to the other losses, the ICI loss provided a better balanced segmentation, and significantly outperformed the Dice loss with an improvement of $1.7-3.7\%$ and the blob loss by $0.6-5.0\%$ in terms of the Dice similarity coefficient on both validation and test set, suggesting that the ICI loss is a potential solution to the instance imbalance problem.
An automated pipeline to create an atlas of in situ hybridization gene expression data in the adult marmoset brain
Mar 13, 2023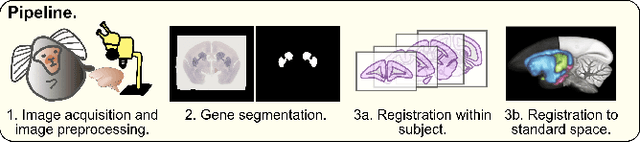
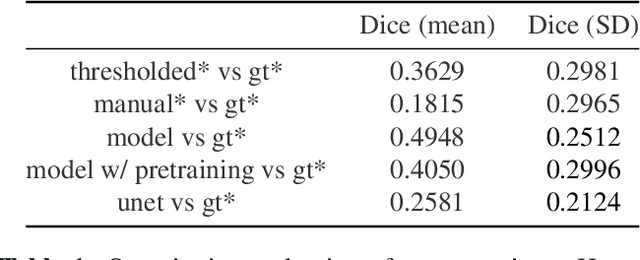
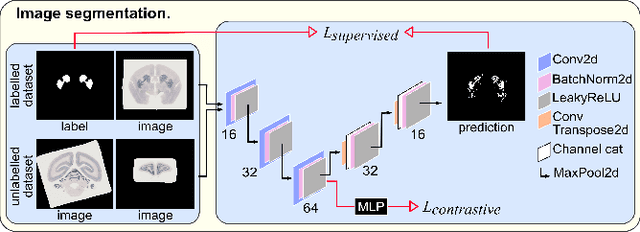

Abstract:We present the first automated pipeline to create an atlas of in situ hybridization gene expression in the adult marmoset brain in the same stereotaxic space. The pipeline consists of segmentation of gene expression from microscopy images and registration of images to a standard space. Automation of this pipeline is necessary to analyze the large volume of data in the genome-wide whole-brain dataset, and to process images that have varying intensity profiles and expression patterns with minimal human bias. To reduce the number of labelled images required for training, we develop a semi-supervised segmentation model. We further develop an iterative algorithm to register images to a standard space, enabling comparative analysis between genes and concurrent visualization with other datasets, thereby facilitating a more holistic understanding of primate brain structure and function.
Deep Learning Convolutional Networks for Multiphoton Microscopy Vasculature Segmentation
Jun 08, 2016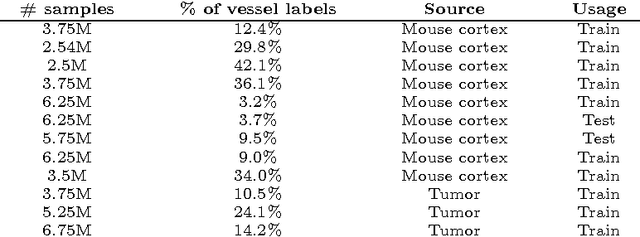
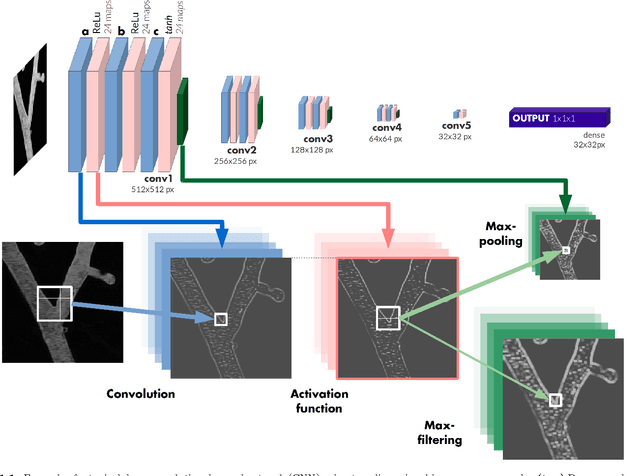
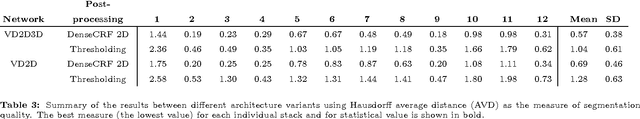
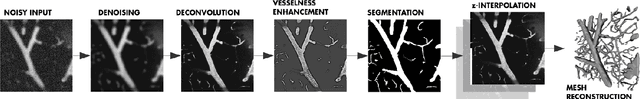
Abstract:Recently there has been an increasing trend to use deep learning frameworks for both 2D consumer images and for 3D medical images. However, there has been little effort to use deep frameworks for volumetric vascular segmentation. We wanted to address this by providing a freely available dataset of 12 annotated two-photon vasculature microscopy stacks. We demonstrated the use of deep learning framework consisting both 2D and 3D convolutional filters (ConvNet). Our hybrid 2D-3D architecture produced promising segmentation result. We derived the architectures from Lee et al. who used the ZNN framework initially designed for electron microscope image segmentation. We hope that by sharing our volumetric vasculature datasets, we will inspire other researchers to experiment with vasculature dataset and improve the used network architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge