Petteri Teikari
Textile IR: A Bidirectional Intermediate Representation for Physics-Aware Fashion CAD
Jan 06, 2026Abstract:We introduce Textile IR, a bidirectional intermediate representation that connects manufacturing-valid CAD, physics-based simulation, and lifecycle assessment for fashion design. Unlike existing siloed tools where pattern software guarantees sewable outputs but understands nothing about drape, and physics simulation predicts behaviour but cannot automatically fix patterns, Textile IR provides the semantic glue for integration through a seven-layer Verification Ladder -- from cheap syntactic checks (pattern closure, seam compatibility) to expensive physics validation (drape simulation, stress analysis). The architecture enables bidirectional feedback: simulation failures suggest pattern modifications; material substitutions update sustainability estimates in real time; uncertainty propagates across the pipeline with explicit confidence bounds. We formalise fashion engineering as constraint satisfaction over three domains and demonstrate how Textile IR's scene-graph representation enables AI systems to manipulate garments as structured programs rather than pixel arrays. The framework addresses the compound uncertainty problem: when measurement errors in material testing, simulation approximations, and LCA database gaps combine, sustainability claims become unreliable without explicit uncertainty tracking. We propose six research priorities and discuss deployment considerations for fashion SMEs where integrated workflows reduce specialised engineering requirements. Key contribution: a formal representation that makes engineering constraints perceptible, manipulable, and immediately consequential -- enabling designers to navigate sustainability, manufacturability, and aesthetic tradeoffs simultaneously rather than discovering conflicts after costly physical prototyping.
Embedded deep learning in ophthalmology: Making ophthalmic imaging smarter
Oct 13, 2018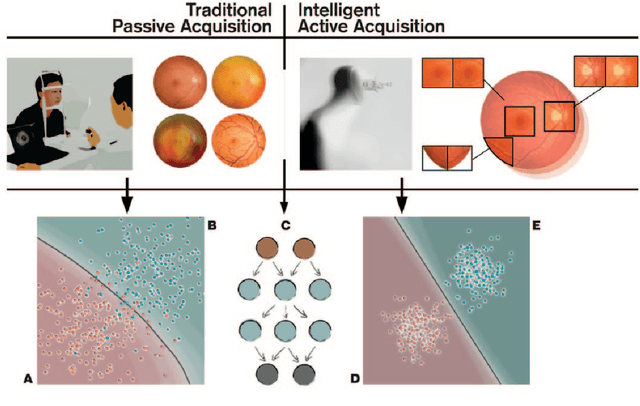
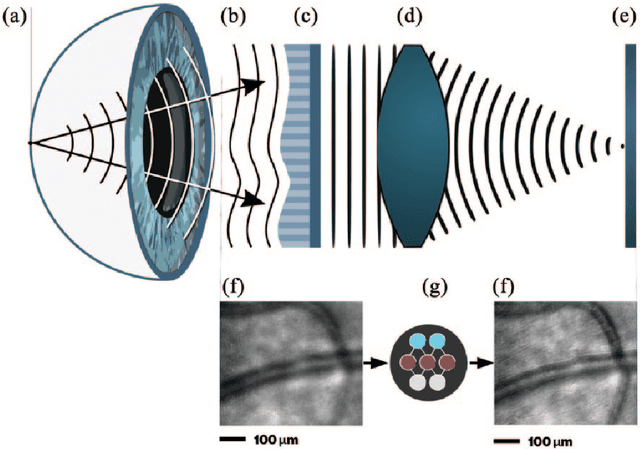
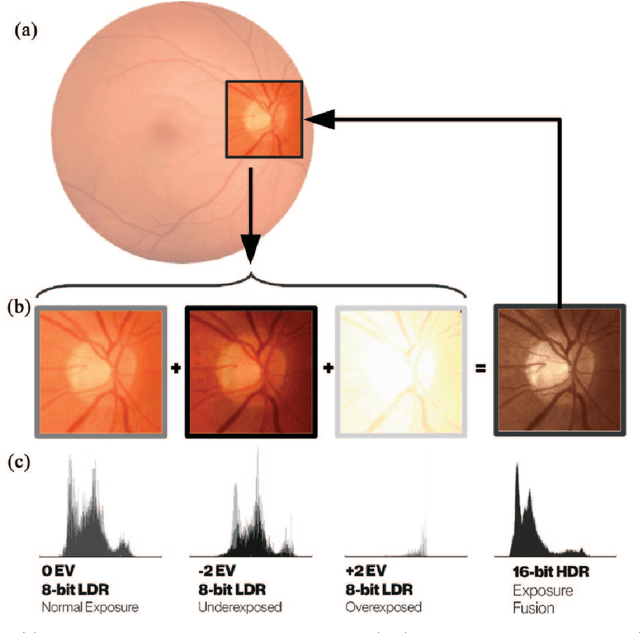
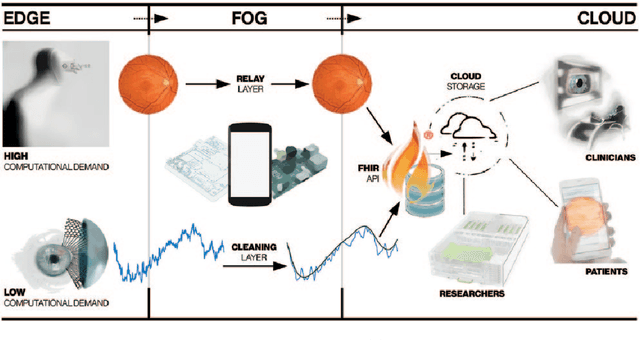
Abstract:Deep learning has recently gained high interest in ophthalmology, due to its ability to detect clinically significant features for diagnosis and prognosis. Despite these significant advances, little is known about the ability of various deep learning systems to be embedded within ophthalmic imaging devices, allowing automated image acquisition. In this work, we will review the existing and future directions for "active acquisition" embedded deep learning, leading to as high quality images with little intervention by the human operator. In clinical practice, the improved image quality should translate into more robust deep learning-based clinical diagnostics. Embedded deep learning will be enabled by the constantly improving hardware performance with low cost. We will briefly review possible computation methods in larger clinical systems. Briefly, they can be included in a three-layer framework composed of edge, fog and cloud layers, the former being performed at a device-level. Improved edge layer performance via "active acquisition" serves as an automatic data curation operator translating to better quality data in electronic health records (EHRs), as well as on the cloud layer, for improved deep learning-based clinical data mining.
Deep Learning Convolutional Networks for Multiphoton Microscopy Vasculature Segmentation
Jun 08, 2016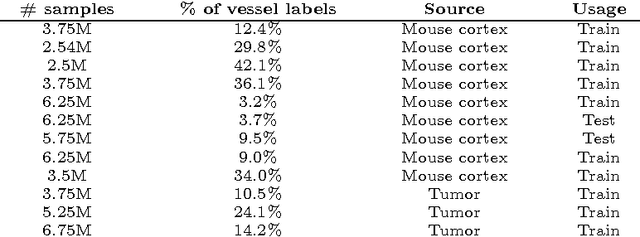
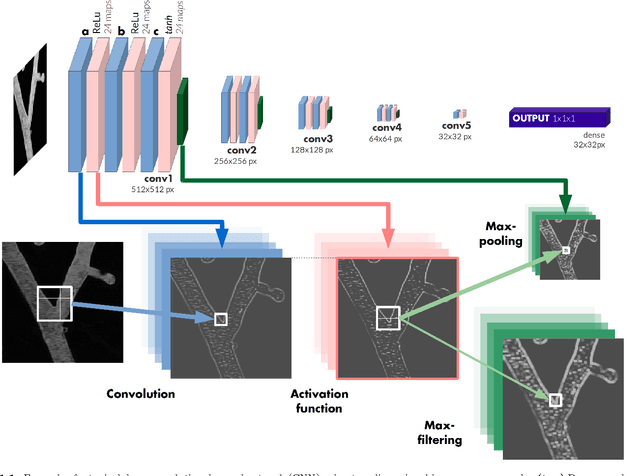
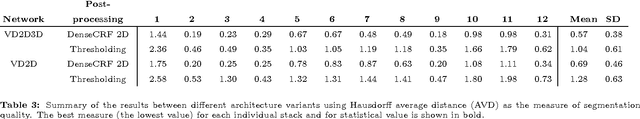
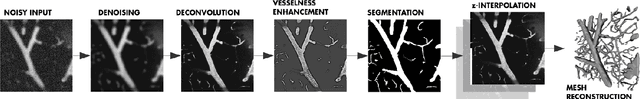
Abstract:Recently there has been an increasing trend to use deep learning frameworks for both 2D consumer images and for 3D medical images. However, there has been little effort to use deep frameworks for volumetric vascular segmentation. We wanted to address this by providing a freely available dataset of 12 annotated two-photon vasculature microscopy stacks. We demonstrated the use of deep learning framework consisting both 2D and 3D convolutional filters (ConvNet). Our hybrid 2D-3D architecture produced promising segmentation result. We derived the architectures from Lee et al. who used the ZNN framework initially designed for electron microscope image segmentation. We hope that by sharing our volumetric vasculature datasets, we will inspire other researchers to experiment with vasculature dataset and improve the used network architectures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge