Raymond P. Najjar
3D Structural Analysis of the Optic Nerve Head to Robustly Discriminate Between Papilledema and Optic Disc Drusen
Dec 18, 2021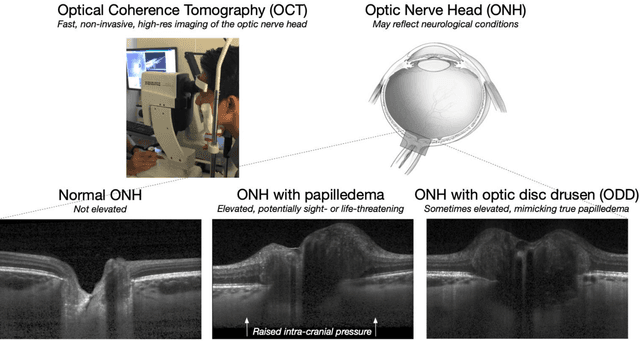
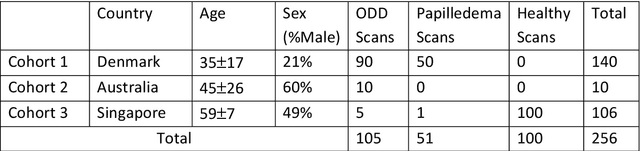
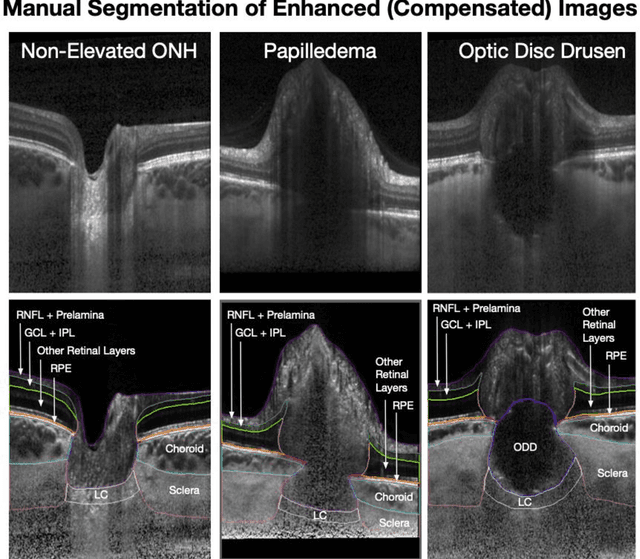
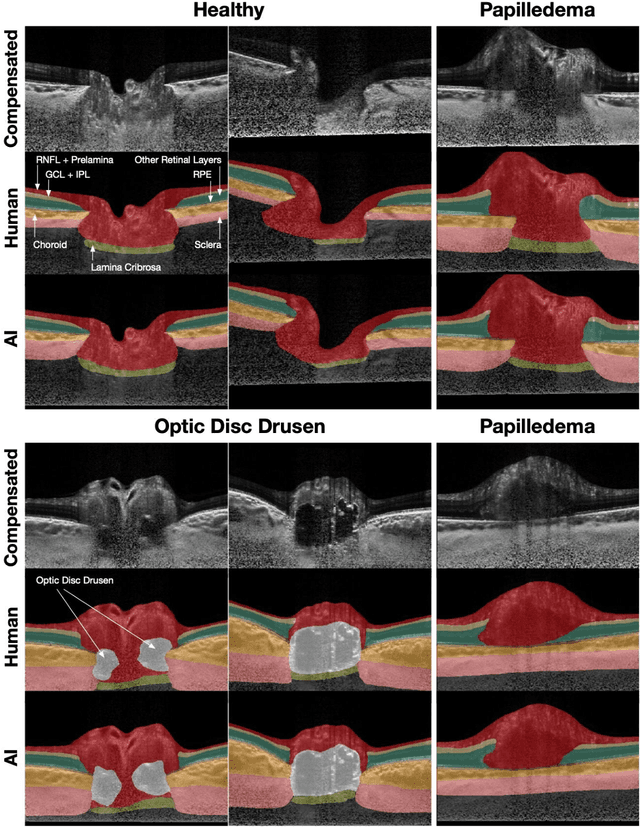
Abstract:Purpose: (1) To develop a deep learning algorithm to identify major tissue structures of the optic nerve head (ONH) in 3D optical coherence tomography (OCT) scans; (2) to exploit such information to robustly differentiate among healthy, optic disc drusen (ODD), and papilledema ONHs. It was a cross-sectional comparative study with confirmed ODD (105 eyes), papilledema due to high intracranial pressure (51 eyes), and healthy controls (100 eyes). 3D scans of the ONHs were acquired using OCT, then processed to improve deep-tissue visibility. At first, a deep learning algorithm was developed using 984 B-scans (from 130 eyes) in order to identify: major neural/connective tissues, and ODD regions. The performance of our algorithm was assessed using the Dice coefficient (DC). In a 2nd step, a classification algorithm (random forest) was designed using 150 OCT volumes to perform 3-class classifications (1: ODD, 2: papilledema, 3: healthy) strictly from their drusen and prelamina swelling scores (derived from the segmentations). To assess performance, we reported the area under the receiver operating characteristic curves (AUCs) for each class. Our segmentation algorithm was able to isolate neural and connective tissues, and ODD regions whenever present. This was confirmed by an average DC of 0.93$\pm$0.03 on the test set, corresponding to good performance. Classification was achieved with high AUCs, i.e. 0.99$\pm$0.01 for the detection of ODD, 0.99 $\pm$ 0.01 for the detection of papilledema, and 0.98$\pm$0.02 for the detection of healthy ONHs. Our AI approach accurately discriminated ODD from papilledema, using a single OCT scan. Our classification performance was excellent, with the caveat that validation in a much larger population is warranted. Our approach may have the potential to establish OCT as the mainstay of diagnostic imaging in neuro-ophthalmology.
Embedded deep learning in ophthalmology: Making ophthalmic imaging smarter
Oct 13, 2018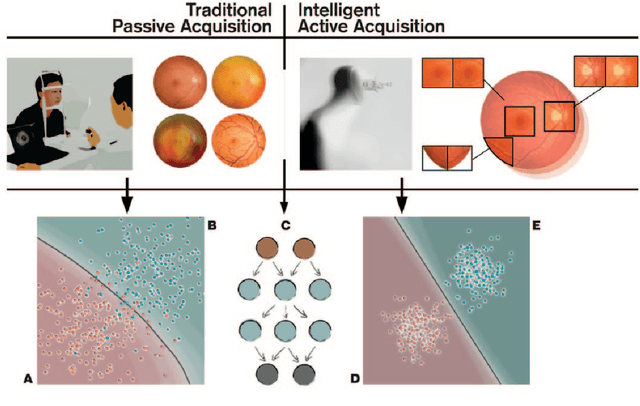
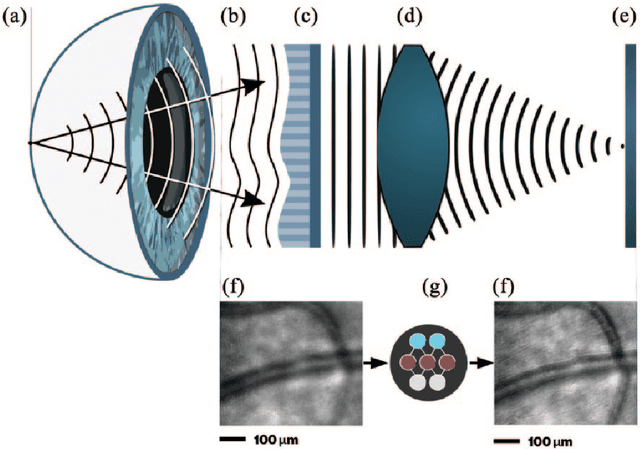
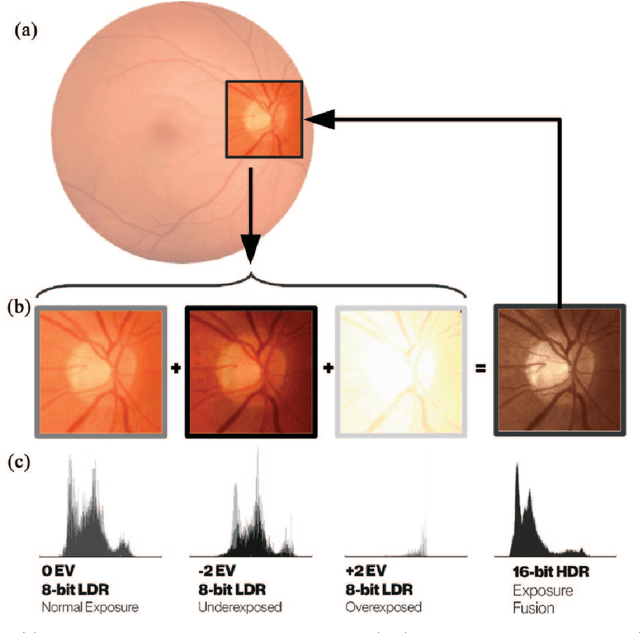
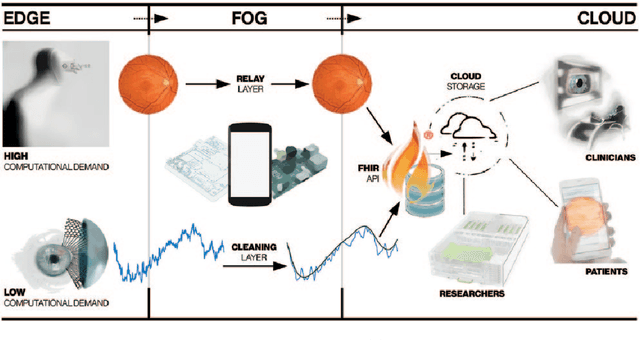
Abstract:Deep learning has recently gained high interest in ophthalmology, due to its ability to detect clinically significant features for diagnosis and prognosis. Despite these significant advances, little is known about the ability of various deep learning systems to be embedded within ophthalmic imaging devices, allowing automated image acquisition. In this work, we will review the existing and future directions for "active acquisition" embedded deep learning, leading to as high quality images with little intervention by the human operator. In clinical practice, the improved image quality should translate into more robust deep learning-based clinical diagnostics. Embedded deep learning will be enabled by the constantly improving hardware performance with low cost. We will briefly review possible computation methods in larger clinical systems. Briefly, they can be included in a three-layer framework composed of edge, fog and cloud layers, the former being performed at a device-level. Improved edge layer performance via "active acquisition" serves as an automatic data curation operator translating to better quality data in electronic health records (EHRs), as well as on the cloud layer, for improved deep learning-based clinical data mining.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge