Michaël J. A. Girard
3D Structural Phenotype of the Optic Nerve Head at the Intersection of Glaucoma and Myopia - A Key to Improving Glaucoma Diagnosis in Myopic Populations
Mar 24, 2025Abstract:Purpose: To characterize the 3D structural phenotypes of the optic nerve head (ONH) in patients with glaucoma, high myopia, and concurrent high myopia and glaucoma, and to evaluate their variations across these conditions. Participants: A total of 685 optical coherence tomography (OCT) scans from 754 subjects of Singapore-Chinese ethnicity, including 256 healthy (H), 94 highly myopic (HM), 227 glaucomatous (G), and 108 highly myopic with glaucoma (HMG) cases. Methods: We segmented the retinal and connective tissues from OCT volumes and their boundary edges were converted into 3D point clouds. To classify the 3D point clouds into four ONH conditions, i.e., H, HM, G, and HMG, a specialized ensemble network was developed, consisting of an encoder to transform high-dimensional input data into a compressed latent vector, a decoder to reconstruct point clouds from the latent vector, and a classifier to categorize the point clouds into the four ONH conditions. Results: The classification network achieved high accuracy, distinguishing H, HM, G, and HMG classes with a micro-average AUC of 0.92 $\pm$ 0.03 on an independent test set. The decoder effectively reconstructed point clouds, achieving a Chamfer loss of 0.013 $\pm$ 0.002. Dimensionality reduction clustered ONHs into four distinct groups, revealing structural variations such as changes in retinal and connective tissue thickness, tilting and stretching of the disc and scleral canal opening, and alterations in optic cup morphology, including shallow or deep excavation, across the four conditions. Conclusions: This study demonstrated that ONHs exhibit distinct structural signatures across H, HM, G, and HMG conditions. The findings further indicate that ONH morphology provides sufficient information for classification into distinct clusters, with principal components capturing unique structural patterns within each group.
Introducing the Biomechanics-Function Relationship in Glaucoma: Improved Visual Field Loss Predictions from intraocular pressure-induced Neural Tissue Strains
Jun 21, 2024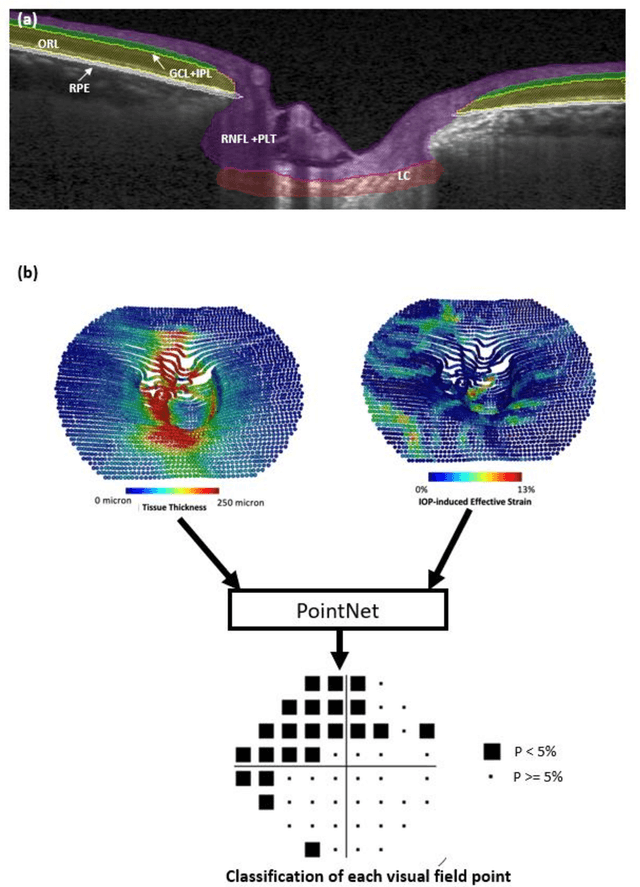

Abstract:Objective. (1) To assess whether neural tissue structure and biomechanics could predict functional loss in glaucoma; (2) To evaluate the importance of biomechanics in making such predictions. Design, Setting and Participants. We recruited 238 glaucoma subjects. For one eye of each subject, we imaged the optic nerve head (ONH) using spectral-domain OCT under the following conditions: (1) primary gaze and (2) primary gaze with acute IOP elevation. Main Outcomes: We utilized automatic segmentation of optic nerve head (ONH) tissues and digital volume correlation (DVC) analysis to compute intraocular pressure (IOP)-induced neural tissue strains. A robust geometric deep learning approach, known as Point-Net, was employed to predict the full Humphrey 24-2 pattern standard deviation (PSD) maps from ONH structural and biomechanical information. For each point in each PSD map, we predicted whether it exhibited no defect or a PSD value of less than 5%. Predictive performance was evaluated using 5-fold cross-validation and the F1-score. We compared the model's performance with and without the inclusion of IOP-induced strains to assess the impact of biomechanics on prediction accuracy. Results: Integrating biomechanical (IOP-induced neural tissue strains) and structural (tissue morphology and neural tissues thickness) information yielded a significantly better predictive model (F1-score: 0.76+-0.02) across validation subjects, as opposed to relying only on structural information, which resulted in a significantly lower F1-score of 0.71+-0.02 (p < 0.05). Conclusion: Our study has shown that the integration of biomechanical data can significantly improve the accuracy of visual field loss predictions. This highlights the importance of the biomechanics-function relationship in glaucoma, and suggests that biomechanics may serve as a crucial indicator for the development and progression of glaucoma.
The 3D Structural Phenotype of the Glaucomatous Optic Nerve Head and its Relationship with The Severity of Visual Field Damage
Jan 07, 2023Abstract:$\bf{Purpose}$: To describe the 3D structural changes in both connective and neural tissues of the optic nerve head (ONH) that occur concurrently at different stages of glaucoma using traditional and AI-driven approaches. $\bf{Methods}$: We included 213 normal, 204 mild glaucoma (mean deviation [MD] $\ge$ -6.00 dB), 118 moderate glaucoma (MD of -6.01 to -12.00 dB), and 118 advanced glaucoma patients (MD < -12.00 dB). All subjects had their ONHs imaged in 3D with Spectralis optical coherence tomography. To describe the 3D structural phenotype of glaucoma as a function of severity, we used two different approaches: (1) We extracted human-defined 3D structural parameters of the ONH including retinal nerve fiber layer (RNFL) thickness, lamina cribrosa (LC) shape and depth at different stages of glaucoma; (2) we also employed a geometric deep learning method (i.e. PointNet) to identify the most important 3D structural features that differentiate ONHs from different glaucoma severity groups without any human input. $\bf{Results}$: We observed that the majority of ONH structural changes occurred in the early glaucoma stage, followed by a plateau effect in the later stages. Using PointNet, we also found that 3D ONH structural changes were present in both neural and connective tissues. In both approaches, we observed that structural changes were more prominent in the superior and inferior quadrant of the ONH, particularly in the RNFL, the prelamina, and the LC. As the severity of glaucoma increased, these changes became more diffuse (i.e. widespread), particularly in the LC. $\bf{Conclusions}$: In this study, we were able to uncover complex 3D structural changes of the ONH in both neural and connective tissues as a function of glaucoma severity. We hope to provide new insights into the complex pathophysiology of glaucoma that might help clinicians in their daily clinical care.
Are Macula or Optic Nerve Head Structures better at Diagnosing Glaucoma? An Answer using AI and Wide-Field Optical Coherence Tomography
Oct 13, 2022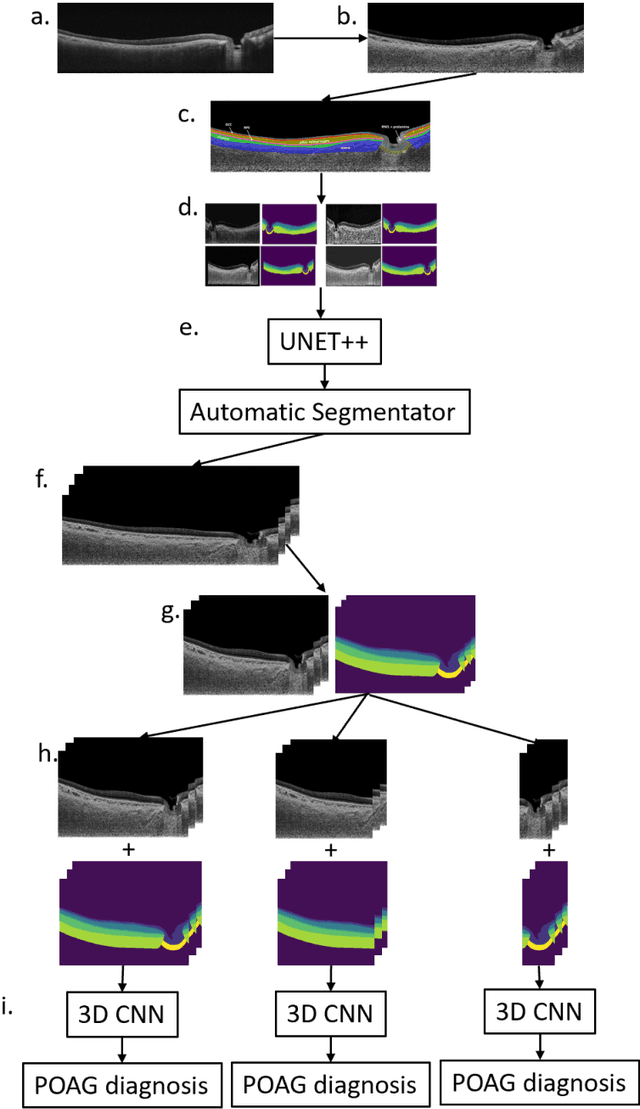
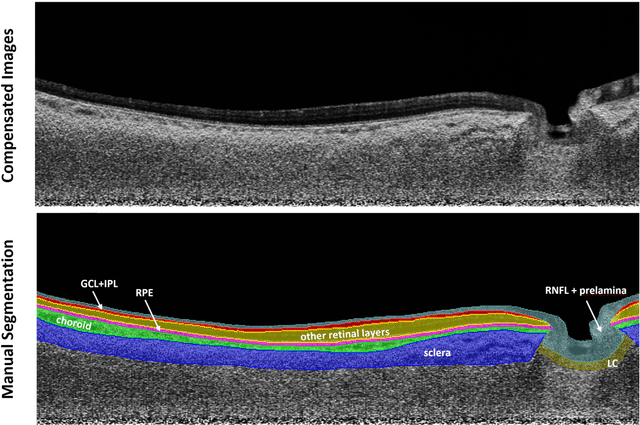
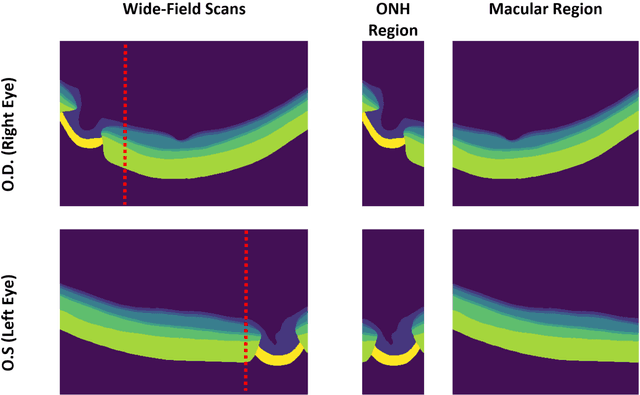
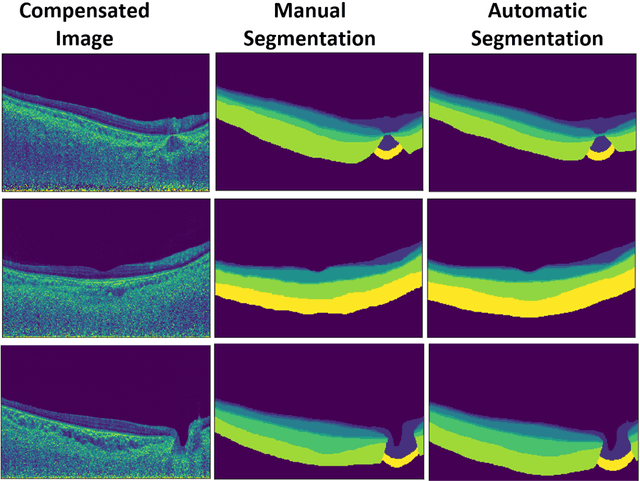
Abstract:Purpose: (1) To develop a deep learning algorithm to automatically segment structures of the optic nerve head (ONH) and macula in 3D wide-field optical coherence tomography (OCT) scans; (2) To assess whether 3D macula or ONH structures (or the combination of both) provide the best diagnostic power for glaucoma. Methods: A cross-sectional comparative study was performed which included wide-field swept-source OCT scans from 319 glaucoma subjects and 298 non-glaucoma subjects. All scans were compensated to improve deep-tissue visibility. We developed a deep learning algorithm to automatically label all major ONH tissue structures by using 270 manually annotated B-scans for training. The performance of our algorithm was assessed using the Dice coefficient (DC). A glaucoma classification algorithm (3D CNN) was then designed using a combination of 500 OCT volumes and their corresponding automatically segmented masks. This algorithm was trained and tested on 3 datasets: OCT scans cropped to contain the macular tissues only, those to contain the ONH tissues only, and the full wide-field OCT scans. The classification performance for each dataset was reported using the AUC. Results: Our segmentation algorithm was able to segment ONH and macular tissues with a DC of 0.94 $\pm$ 0.003. The classification algorithm was best able to diagnose glaucoma using wide-field 3D-OCT volumes with an AUC of 0.99 $\pm$ 0.01, followed by ONH volumes with an AUC of 0.93 $\pm$ 0.06, and finally macular volumes with an AUC of 0.91 $\pm$ 0.11. Conclusions: this study showed that using wide-field OCT as compared to the typical OCT images containing just the ONH or macular may allow for a significantly improved glaucoma diagnosis. This may encourage the mainstream adoption of 3D wide-field OCT scans. For clinical AI studies that use traditional machines, we would recommend the use of ONH scans as opposed to macula scans.
AI-based Clinical Assessment of Optic Nerve Head Robustness Superseding Biomechanical Testing
Jun 09, 2022
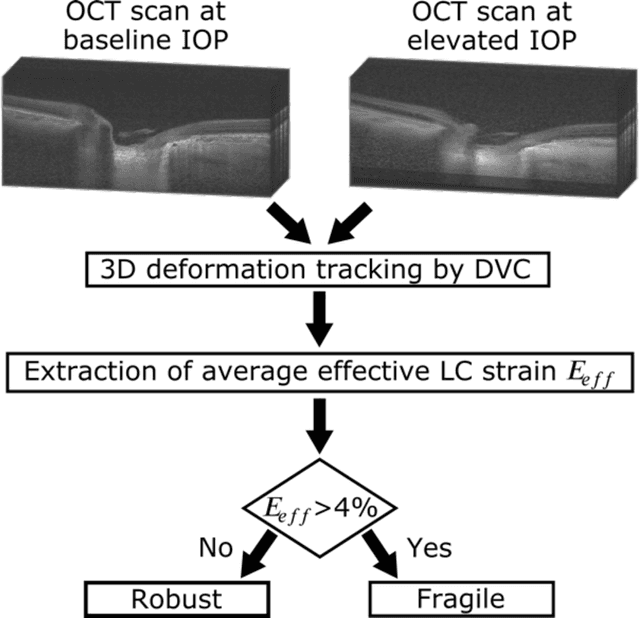
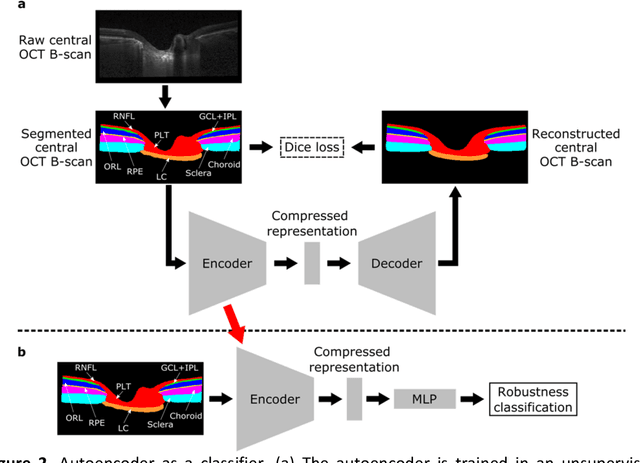
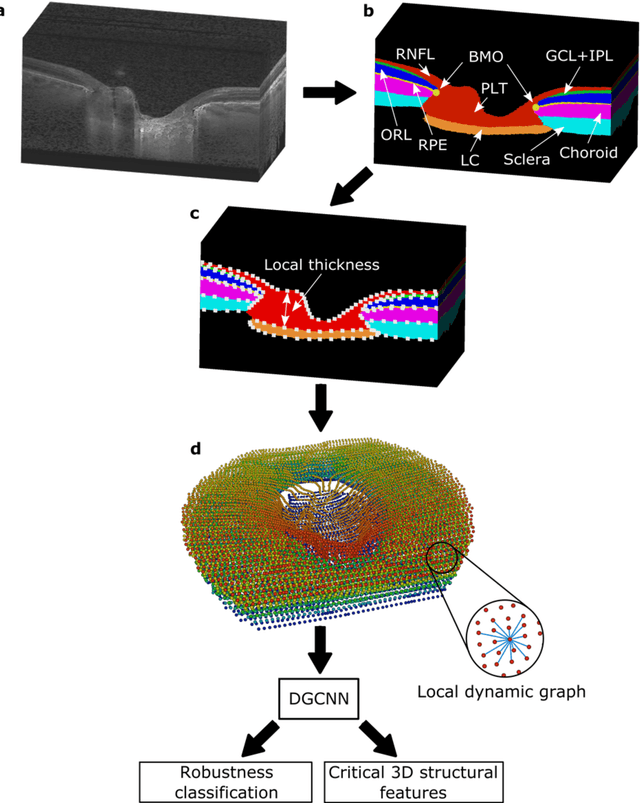
Abstract:$\mathbf{Purpose}$: To use artificial intelligence (AI) to: (1) exploit biomechanical knowledge of the optic nerve head (ONH) from a relatively large population; (2) assess ONH robustness from a single optical coherence tomography (OCT) scan of the ONH; (3) identify what critical three-dimensional (3D) structural features make a given ONH robust. $\mathbf{Design}$: Retrospective cross-sectional study. $\mathbf{Methods}$: 316 subjects had their ONHs imaged with OCT before and after acute intraocular pressure (IOP) elevation through ophthalmo-dynamometry. IOP-induced lamina-cribrosa deformations were then mapped in 3D and used to classify ONHs. Those with LC deformations superior to 4% were considered fragile, while those with deformations inferior to 4% robust. Learning from these data, we compared three AI algorithms to predict ONH robustness strictly from a baseline (undeformed) OCT volume: (1) a random forest classifier; (2) an autoencoder; and (3) a dynamic graph CNN (DGCNN). The latter algorithm also allowed us to identify what critical 3D structural features make a given ONH robust. $\mathbf{Results}$: All 3 methods were able to predict ONH robustness from 3D structural information alone and without the need to perform biomechanical testing. The DGCNN (area under the receiver operating curve [AUC]: 0.76 $\pm$ 0.08) outperformed the autoencoder (AUC: 0.70 $\pm$ 0.07) and the random forest classifier (AUC: 0.69 $\pm$ 0.05). Interestingly, to assess ONH robustness, the DGCNN mainly used information from the scleral canal and the LC insertion sites. $\mathbf{Conclusions}$: We propose an AI-driven approach that can assess the robustness of a given ONH solely from a single OCT scan of the ONH, and without the need to perform biomechanical testing. Longitudinal studies should establish whether ONH robustness could help us identify fast visual field loss progressors.
Geometric Deep Learning to Identify the Critical 3D Structural Features of the Optic Nerve Head for Glaucoma Diagnosis
Apr 20, 2022

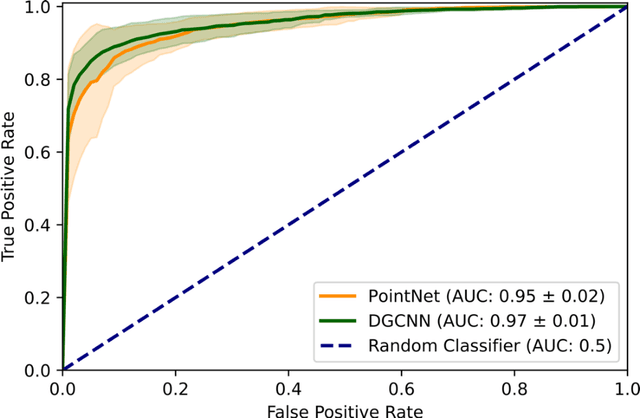

Abstract:Purpose: The optic nerve head (ONH) undergoes complex and deep 3D morphological changes during the development and progression of glaucoma. Optical coherence tomography (OCT) is the current gold standard to visualize and quantify these changes, however the resulting 3D deep-tissue information has not yet been fully exploited for the diagnosis and prognosis of glaucoma. To this end, we aimed: (1) To compare the performance of two relatively recent geometric deep learning techniques in diagnosing glaucoma from a single OCT scan of the ONH; and (2) To identify the 3D structural features of the ONH that are critical for the diagnosis of glaucoma. Methods: In this study, we included a total of 2,247 non-glaucoma and 2,259 glaucoma scans from 1,725 subjects. All subjects had their ONHs imaged in 3D with Spectralis OCT. All OCT scans were automatically segmented using deep learning to identify major neural and connective tissues. Each ONH was then represented as a 3D point cloud. We used PointNet and dynamic graph convolutional neural network (DGCNN) to diagnose glaucoma from such 3D ONH point clouds and to identify the critical 3D structural features of the ONH for glaucoma diagnosis. Results: Both the DGCNN (AUC: 0.97$\pm$0.01) and PointNet (AUC: 0.95$\pm$0.02) were able to accurately detect glaucoma from 3D ONH point clouds. The critical points formed an hourglass pattern with most of them located in the inferior and superior quadrant of the ONH. Discussion: The diagnostic accuracy of both geometric deep learning approaches was excellent. Moreover, we were able to identify the critical 3D structural features of the ONH for glaucoma diagnosis that tremendously improved the transparency and interpretability of our method. Consequently, our approach may have strong potential to be used in clinical applications for the diagnosis and prognosis of a wide range of ophthalmic disorders.
3D Structural Analysis of the Optic Nerve Head to Robustly Discriminate Between Papilledema and Optic Disc Drusen
Dec 18, 2021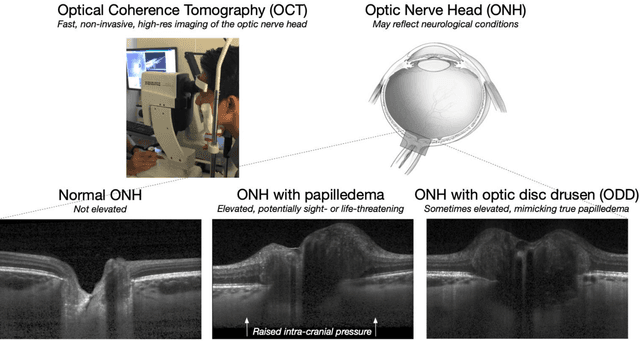
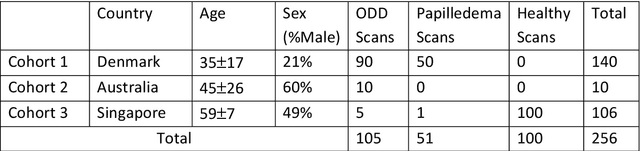
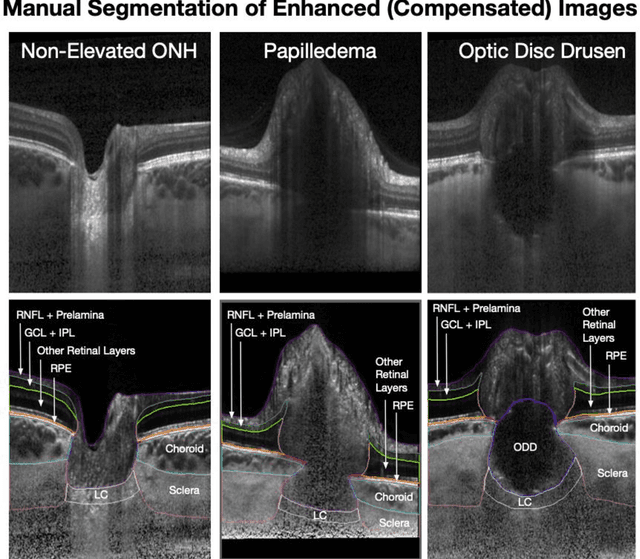
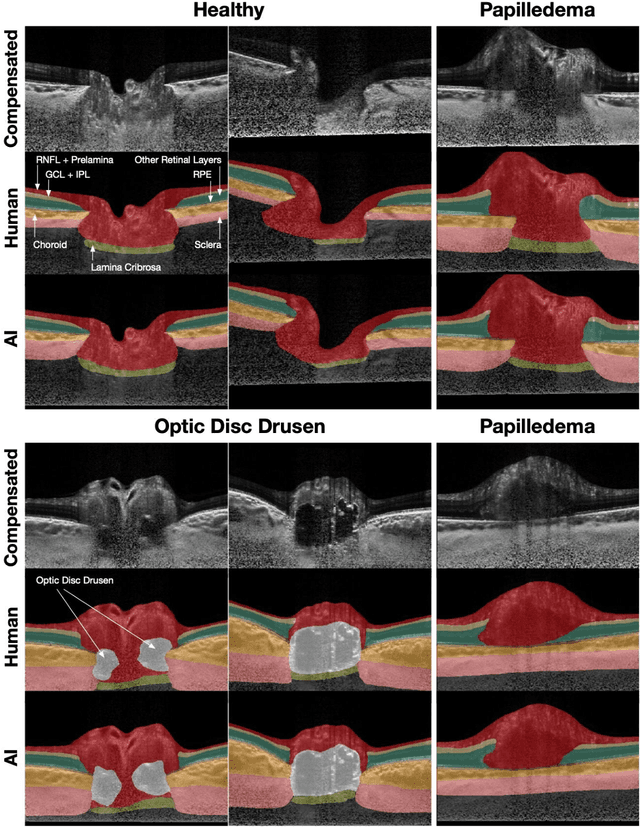
Abstract:Purpose: (1) To develop a deep learning algorithm to identify major tissue structures of the optic nerve head (ONH) in 3D optical coherence tomography (OCT) scans; (2) to exploit such information to robustly differentiate among healthy, optic disc drusen (ODD), and papilledema ONHs. It was a cross-sectional comparative study with confirmed ODD (105 eyes), papilledema due to high intracranial pressure (51 eyes), and healthy controls (100 eyes). 3D scans of the ONHs were acquired using OCT, then processed to improve deep-tissue visibility. At first, a deep learning algorithm was developed using 984 B-scans (from 130 eyes) in order to identify: major neural/connective tissues, and ODD regions. The performance of our algorithm was assessed using the Dice coefficient (DC). In a 2nd step, a classification algorithm (random forest) was designed using 150 OCT volumes to perform 3-class classifications (1: ODD, 2: papilledema, 3: healthy) strictly from their drusen and prelamina swelling scores (derived from the segmentations). To assess performance, we reported the area under the receiver operating characteristic curves (AUCs) for each class. Our segmentation algorithm was able to isolate neural and connective tissues, and ODD regions whenever present. This was confirmed by an average DC of 0.93$\pm$0.03 on the test set, corresponding to good performance. Classification was achieved with high AUCs, i.e. 0.99$\pm$0.01 for the detection of ODD, 0.99 $\pm$ 0.01 for the detection of papilledema, and 0.98$\pm$0.02 for the detection of healthy ONHs. Our AI approach accurately discriminated ODD from papilledema, using a single OCT scan. Our classification performance was excellent, with the caveat that validation in a much larger population is warranted. Our approach may have the potential to establish OCT as the mainstay of diagnostic imaging in neuro-ophthalmology.
Describing the Structural Phenotype of the Glaucomatous Optic Nerve Head Using Artificial Intelligence
Dec 17, 2020
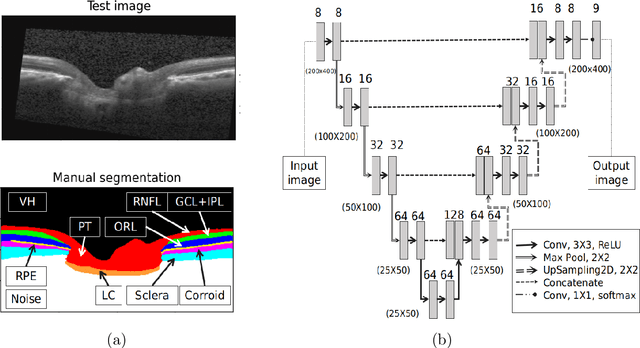
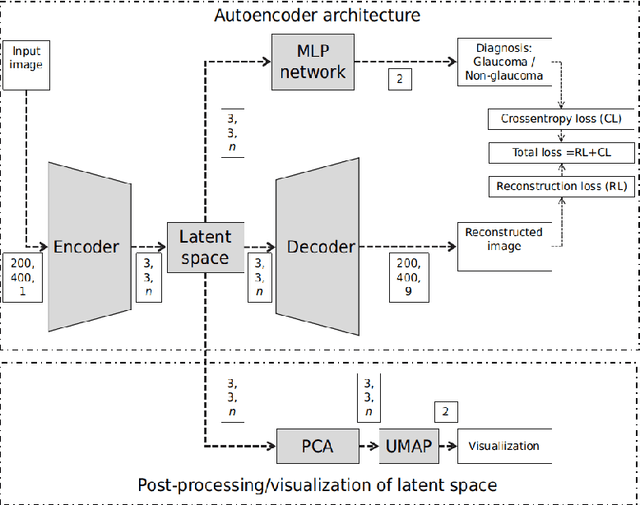

Abstract:The optic nerve head (ONH) typically experiences complex neural- and connective-tissue structural changes with the development and progression of glaucoma, and monitoring these changes could be critical for improved diagnosis and prognosis in the glaucoma clinic. The gold-standard technique to assess structural changes of the ONH clinically is optical coherence tomography (OCT). However, OCT is limited to the measurement of a few hand-engineered parameters, such as the thickness of the retinal nerve fiber layer (RNFL), and has not yet been qualified as a stand-alone device for glaucoma diagnosis and prognosis applications. We argue this is because the vast amount of information available in a 3D OCT scan of the ONH has not been fully exploited. In this study we propose a deep learning approach that can: \textbf{(1)} fully exploit information from an OCT scan of the ONH; \textbf{(2)} describe the structural phenotype of the glaucomatous ONH; and that can \textbf{(3)} be used as a robust glaucoma diagnosis tool. Specifically, the structural features identified by our algorithm were found to be related to clinical observations of glaucoma. The diagnostic accuracy from these structural features was $92.0 \pm 2.3 \%$ with a sensitivity of $90.0 \pm 2.4 \% $ (at $95 \%$ specificity). By changing their magnitudes in steps, we were able to reveal how the morphology of the ONH changes as one transitions from a `non-glaucoma' to a `glaucoma' condition. We believe our work may have strong clinical implication for our understanding of glaucoma pathogenesis, and could be improved in the future to also predict future loss of vision.
OCT-GAN: Single Step Shadow and Noise Removal from Optical Coherence Tomography Images of the Human Optic Nerve Head
Oct 06, 2020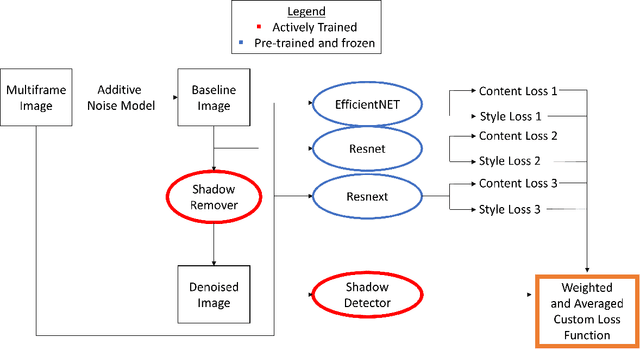
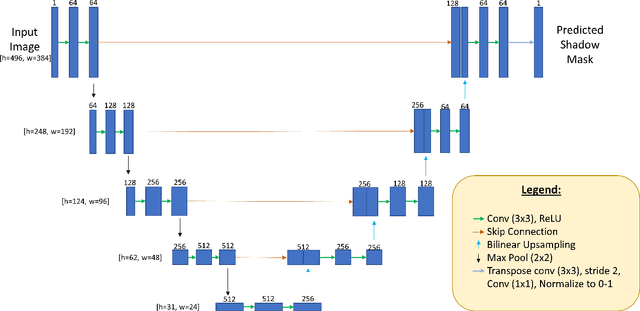
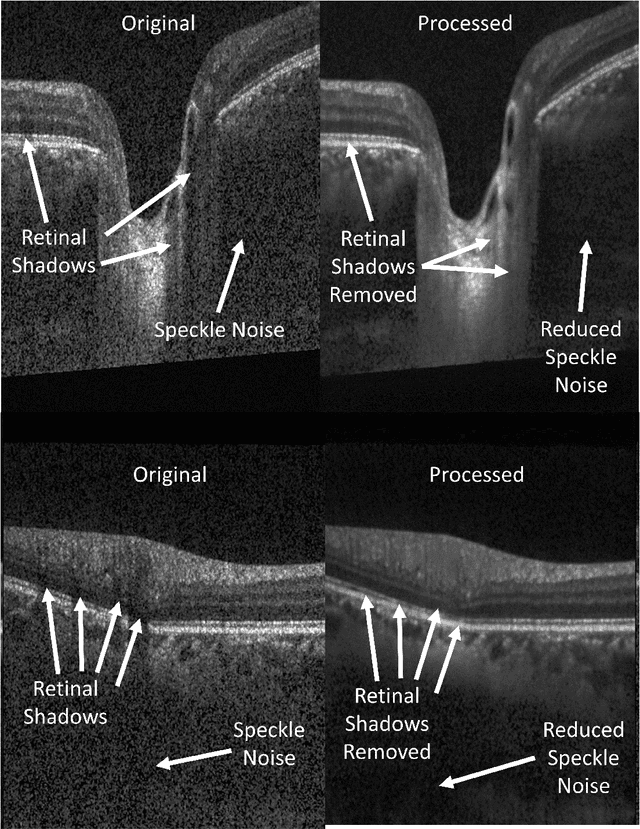
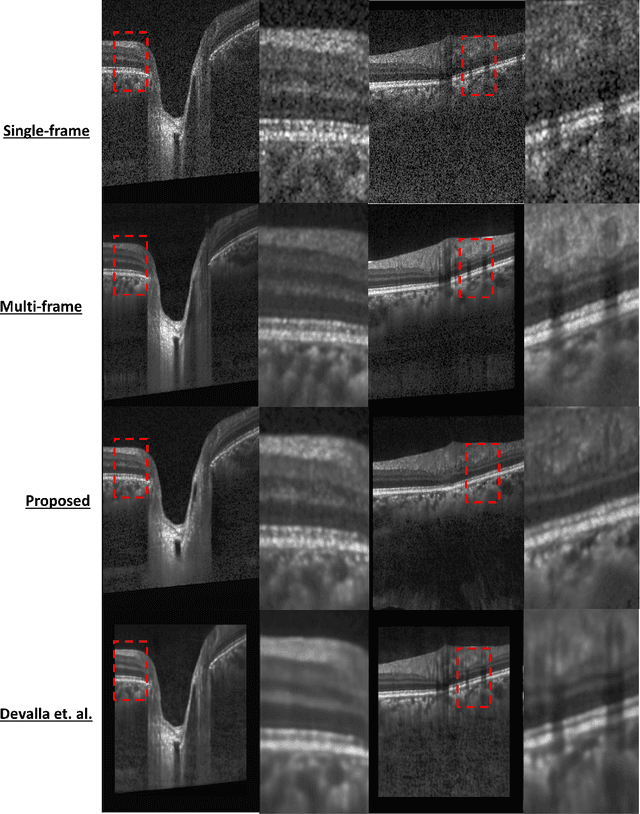
Abstract:Speckle noise and retinal shadows within OCT B-scans occlude important edges, fine textures and deep tissues, preventing accurate and robust diagnosis by algorithms and clinicians. We developed a single process that successfully removed both noise and retinal shadows from unseen single-frame B-scans within 10.4ms. Mean average gradient magnitude (AGM) for the proposed algorithm was 57.2% higher than current state-of-the-art, while mean peak signal to noise ratio (PSNR), contrast to noise ratio (CNR), and structural similarity index metric (SSIM) increased by 11.1%, 154% and 187% respectively compared to single-frame B-scans. Mean intralayer contrast (ILC) improvement for the retinal nerve fiber layer (RNFL), photoreceptor layer (PR) and retinal pigment epithelium (RPE) layers decreased from 0.362 \pm 0.133 to 0.142 \pm 0.102, 0.449 \pm 0.116 to 0.0904 \pm 0.0769, 0.381 \pm 0.100 to 0.0590 \pm 0.0451 respectively. The proposed algorithm reduces the necessity for long image acquisition times, minimizes expensive hardware requirements and reduces motion artifacts in OCT images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge