Swati Sharma
3D Structural Phenotype of the Optic Nerve Head at the Intersection of Glaucoma and Myopia - A Key to Improving Glaucoma Diagnosis in Myopic Populations
Mar 24, 2025Abstract:Purpose: To characterize the 3D structural phenotypes of the optic nerve head (ONH) in patients with glaucoma, high myopia, and concurrent high myopia and glaucoma, and to evaluate their variations across these conditions. Participants: A total of 685 optical coherence tomography (OCT) scans from 754 subjects of Singapore-Chinese ethnicity, including 256 healthy (H), 94 highly myopic (HM), 227 glaucomatous (G), and 108 highly myopic with glaucoma (HMG) cases. Methods: We segmented the retinal and connective tissues from OCT volumes and their boundary edges were converted into 3D point clouds. To classify the 3D point clouds into four ONH conditions, i.e., H, HM, G, and HMG, a specialized ensemble network was developed, consisting of an encoder to transform high-dimensional input data into a compressed latent vector, a decoder to reconstruct point clouds from the latent vector, and a classifier to categorize the point clouds into the four ONH conditions. Results: The classification network achieved high accuracy, distinguishing H, HM, G, and HMG classes with a micro-average AUC of 0.92 $\pm$ 0.03 on an independent test set. The decoder effectively reconstructed point clouds, achieving a Chamfer loss of 0.013 $\pm$ 0.002. Dimensionality reduction clustered ONHs into four distinct groups, revealing structural variations such as changes in retinal and connective tissue thickness, tilting and stretching of the disc and scleral canal opening, and alterations in optic cup morphology, including shallow or deep excavation, across the four conditions. Conclusions: This study demonstrated that ONHs exhibit distinct structural signatures across H, HM, G, and HMG conditions. The findings further indicate that ONH morphology provides sufficient information for classification into distinct clusters, with principal components capturing unique structural patterns within each group.
Enabling Adoption of Regenerative Agriculture through Soil Carbon Copilots
Nov 25, 2024



Abstract:Mitigating climate change requires transforming agriculture to minimize environ mental impact and build climate resilience. Regenerative agricultural practices enhance soil organic carbon (SOC) levels, thus improving soil health and sequestering carbon. A challenge to increasing regenerative agriculture practices is cheaply measuring SOC over time and understanding how SOC is affected by regenerative agricultural practices and other environmental factors and farm management practices. To address this challenge, we introduce an AI-driven Soil Organic Carbon Copilot that automates the ingestion of complex multi-resolution, multi-modal data to provide large-scale insights into soil health and regenerative practices. Our data includes extreme weather event data (e.g., drought and wildfire incidents), farm management data (e.g., cropland information and tillage predictions), and SOC predictions. We find that integrating public data and specialized models enables large-scale, localized analysis for sustainable agriculture. In comparisons of agricultural practices across California counties, we find evidence that diverse agricultural activity may mitigate the negative effects of tillage; and that while extreme weather conditions heavily affect SOC, composting may mitigate SOC loss. Finally, implementing role-specific personas empowers agronomists, farm consultants, policymakers, and other stakeholders to implement evidence-based strategies that promote sustainable agriculture and build climate resilience.
RAG vs Fine-tuning: Pipelines, Tradeoffs, and a Case Study on Agriculture
Jan 30, 2024Abstract:There are two common ways in which developers are incorporating proprietary and domain-specific data when building applications of Large Language Models (LLMs): Retrieval-Augmented Generation (RAG) and Fine-Tuning. RAG augments the prompt with the external data, while fine-Tuning incorporates the additional knowledge into the model itself. However, the pros and cons of both approaches are not well understood. In this paper, we propose a pipeline for fine-tuning and RAG, and present the tradeoffs of both for multiple popular LLMs, including Llama2-13B, GPT-3.5, and GPT-4. Our pipeline consists of multiple stages, including extracting information from PDFs, generating questions and answers, using them for fine-tuning, and leveraging GPT-4 for evaluating the results. We propose metrics to assess the performance of different stages of the RAG and fine-Tuning pipeline. We conduct an in-depth study on an agricultural dataset. Agriculture as an industry has not seen much penetration of AI, and we study a potentially disruptive application - what if we could provide location-specific insights to a farmer? Our results show the effectiveness of our dataset generation pipeline in capturing geographic-specific knowledge, and the quantitative and qualitative benefits of RAG and fine-tuning. We see an accuracy increase of over 6 p.p. when fine-tuning the model and this is cumulative with RAG, which increases accuracy by 5 p.p. further. In one particular experiment, we also demonstrate that the fine-tuned model leverages information from across geographies to answer specific questions, increasing answer similarity from 47% to 72%. Overall, the results point to how systems built using LLMs can be adapted to respond and incorporate knowledge across a dimension that is critical for a specific industry, paving the way for further applications of LLMs in other industrial domains.
Domain Adaptation for Sustainable Soil Management using Causal and Contrastive Constraint Minimization
Jan 13, 2024Abstract:Monitoring organic matter is pivotal for maintaining soil health and can help inform sustainable soil management practices. While sensor-based soil information offers higher-fidelity and reliable insights into organic matter changes, sampling and measuring sensor data is cost-prohibitive. We propose a multi-modal, scalable framework that can estimate organic matter from remote sensing data, a more readily available data source while leveraging sparse soil information for improving generalization. Using the sensor data, we preserve underlying causal relations among sensor attributes and organic matter. Simultaneously we leverage inherent structure in the data and train the model to discriminate among domains using contrastive learning. This causal and contrastive constraint minimization ensures improved generalization and adaptation to other domains. We also shed light on the interpretability of the framework by identifying attributes that are important for improving generalization. Identifying these key soil attributes that affect organic matter will aid in efforts to standardize data collection efforts.
Knowledge Guided Representation Learning and Causal Structure Learning in Soil Science
Jun 15, 2023



Abstract:An improved understanding of soil can enable more sustainable land-use practices. Nevertheless, soil is called a complex, living medium due to the complex interaction of different soil processes that limit our understanding of soil. Process-based models and analyzing observed data provide two avenues for improving our understanding of soil processes. Collecting observed data is cost-prohibitive but reflects real-world behavior, while process-based models can be used to generate ample synthetic data which may not be representative of reality. We propose a framework, knowledge-guided representation learning, and causal structure learning (KGRCL), to accelerate scientific discoveries in soil science. The framework improves representation learning for simulated soil processes via conditional distribution matching with observed soil processes. Simultaneously, the framework leverages both observed and simulated data to learn a causal structure among the soil processes. The learned causal graph is more representative of ground truth than other graphs generated from other causal discovery methods. Furthermore, the learned causal graph is leveraged in a supervised learning setup to predict the impact of fertilizer use and changing weather on soil carbon. We present the results in five different locations to show the improvement in the prediction performance in out-of-sample and few-shots setting.
DeepG2P: Fusing Multi-Modal Data to Improve Crop Production
Nov 11, 2022



Abstract:Agriculture is at the heart of the solution to achieve sustainability in feeding the world population, but advancing our understanding on how agricultural output responds to climatic variability is still needed. Precision Agriculture (PA), which is a management strategy that uses technology such as remote sensing, Geographical Information System (GIS), and machine learning for decision making in the field, has emerged as a promising approach to enhance crop production, increase yield, and reduce water and nutrient losses and environmental impacts. In this context, multiple models to predict agricultural phenotypes, such as crop yield, from genomics (G), environment (E), weather and soil, and field management practices (M) have been developed. These models have traditionally been based on mechanistic or statistical approaches. However, AI approaches are intrinsically well-suited to model complex interactions and have more recently been developed, outperforming classical methods. Here, we present a Natural Language Processing (NLP)-based neural network architecture to process the G, E and M inputs and their interactions. We show that by modeling DNA as natural language, our approach performs better than previous approaches when tested for new environments and similarly to other approaches for unseen seed varieties.
Causal Modeling of Soil Processes for Improved Generalization
Nov 10, 2022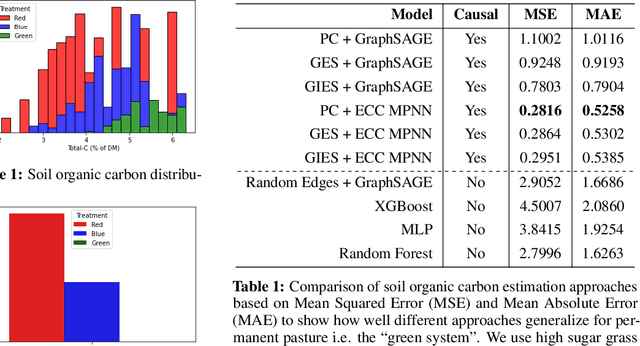
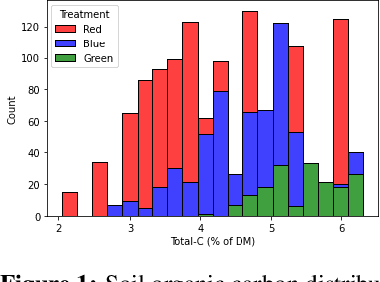
Abstract:Measuring and monitoring soil organic carbon is critical for agricultural productivity and for addressing critical environmental problems. Soil organic carbon not only enriches nutrition in soil, but also has a gamut of co-benefits such as improving water storage and limiting physical erosion. Despite a litany of work in soil organic carbon estimation, current approaches do not generalize well across soil conditions and management practices. We empirically show that explicit modeling of cause-and-effect relationships among the soil processes improves the out-of-distribution generalizability of prediction models. We provide a comparative analysis of soil organic carbon estimation models where the skeleton is estimated using causal discovery methods. Our framework provide an average improvement of 81% in test mean squared error and 52% in test mean absolute error.
Machine learning can guide experimental approaches for protein digestibility estimations
Nov 01, 2022



Abstract:Food protein digestibility and bioavailability are critical aspects in addressing human nutritional demands, particularly when seeking sustainable alternatives to animal-based proteins. In this study, we propose a machine learning approach to predict the true ileal digestibility coefficient of food items. The model makes use of a unique curated dataset that combines nutritional information from different foods with FASTA sequences of some of their protein families. We extracted the biochemical properties of the proteins and combined these properties with embeddings from a Transformer-based protein Language Model (pLM). In addition, we used SHAP to identify features that contribute most to the model prediction and provide interpretability. This first AI-based model for predicting food protein digestibility has an accuracy of 90% compared to existing experimental techniques. With this accuracy, our model can eliminate the need for lengthy in-vivo or in-vitro experiments, making the process of creating new foods faster, cheaper, and more ethical.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge