Sripad Krishna Devalla
Towards A Device-Independent Deep Learning Approach for the Automated Segmentation of Sonographic Fetal Brain Structures: A Multi-Center and Multi-Device Validation
Feb 28, 2022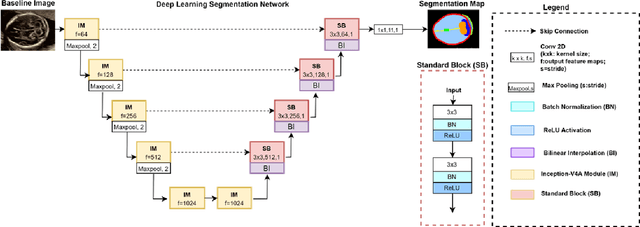
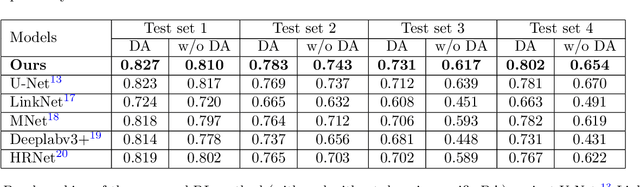
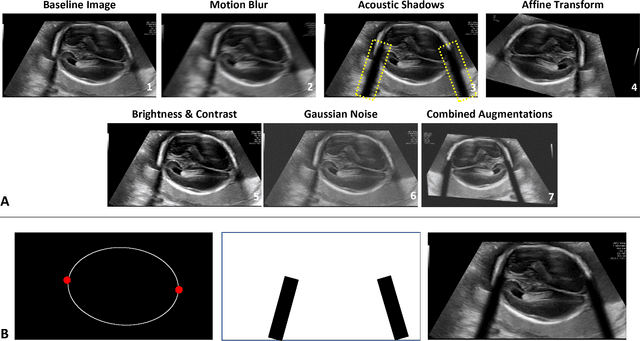
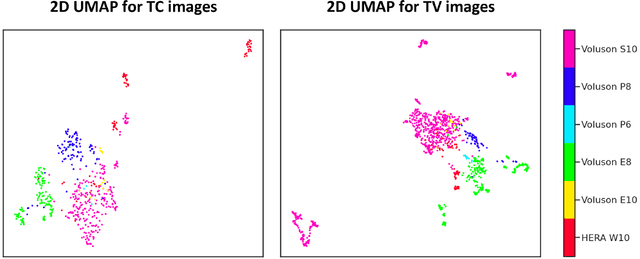
Abstract:Quality assessment of prenatal ultrasonography is essential for the screening of fetal central nervous system (CNS) anomalies. The interpretation of fetal brain structures is highly subjective, expertise-driven, and requires years of training experience, limiting quality prenatal care for all pregnant mothers. With recent advancement in Artificial Intelligence (AI), specifically deep learning (DL), assistance in precise anatomy identification through semantic segmentation essential for the reliable assessment of growth and neurodevelopment, and detection of structural abnormalities have been proposed. However, existing works only identify certain structures (e.g., cavum septum pellucidum, lateral ventricles, cerebellum) from either of the axial views (transventricular, transcerebellar), limiting the scope for a thorough anatomical assessment as per practice guidelines necessary for the screening of CNS anomalies. Further, existing works do not analyze the generalizability of these DL algorithms across images from multiple ultrasound devices and centers, thus, limiting their real-world clinical impact. In this study, we propose a DL based segmentation framework for the automated segmentation of 10 key fetal brain structures from 2 axial planes from fetal brain USG images (2D). We developed a custom U-Net variant that uses inceptionv4 block as a feature extractor and leverages custom domain-specific data augmentation. Quantitatively, the mean (10 structures; test sets 1/2/3/4) Dice-coefficients were: 0.827, 0.802, 0.731, 0.783. Irrespective of the USG device/center, the DL segmentations were qualitatively comparable to their manual segmentations. The proposed DL system offered a promising and generalizable performance (multi-centers, multi-device) and also presents evidence in support of device-induced variation in image quality (a challenge to generalizibility) by using UMAP analysis.
OCT-GAN: Single Step Shadow and Noise Removal from Optical Coherence Tomography Images of the Human Optic Nerve Head
Oct 06, 2020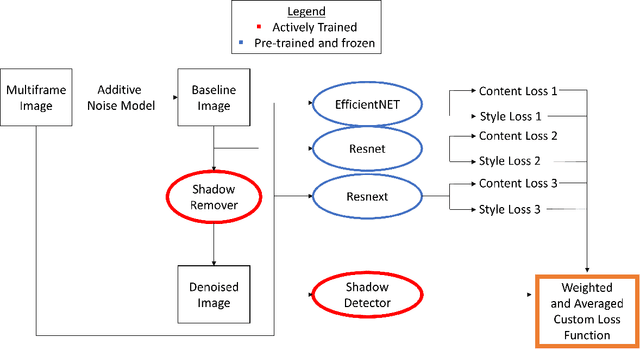
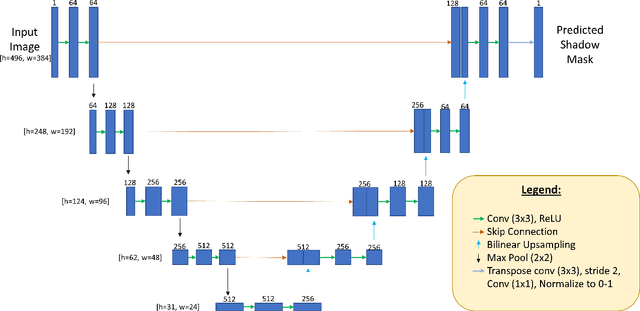
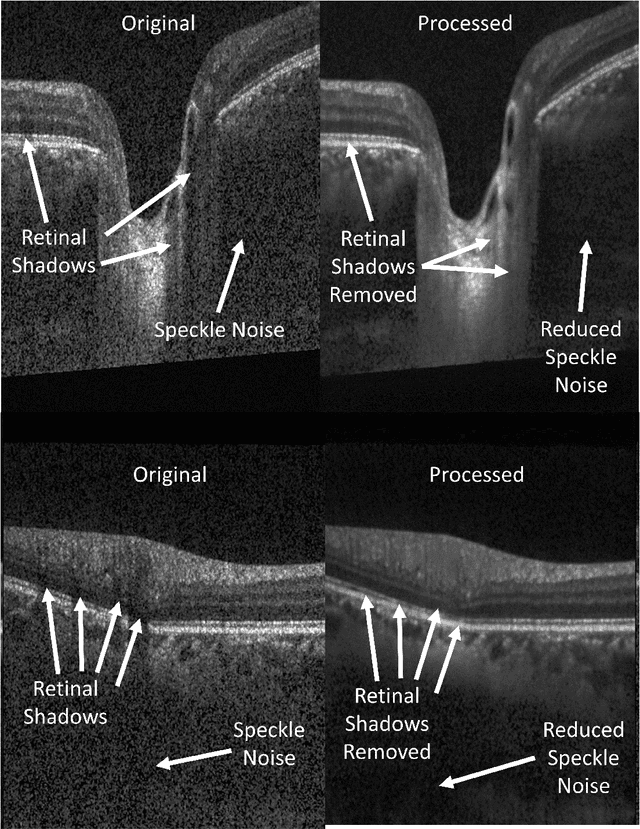
Abstract:Speckle noise and retinal shadows within OCT B-scans occlude important edges, fine textures and deep tissues, preventing accurate and robust diagnosis by algorithms and clinicians. We developed a single process that successfully removed both noise and retinal shadows from unseen single-frame B-scans within 10.4ms. Mean average gradient magnitude (AGM) for the proposed algorithm was 57.2% higher than current state-of-the-art, while mean peak signal to noise ratio (PSNR), contrast to noise ratio (CNR), and structural similarity index metric (SSIM) increased by 11.1%, 154% and 187% respectively compared to single-frame B-scans. Mean intralayer contrast (ILC) improvement for the retinal nerve fiber layer (RNFL), photoreceptor layer (PR) and retinal pigment epithelium (RPE) layers decreased from 0.362 \pm 0.133 to 0.142 \pm 0.102, 0.449 \pm 0.116 to 0.0904 \pm 0.0769, 0.381 \pm 0.100 to 0.0590 \pm 0.0451 respectively. The proposed algorithm reduces the necessity for long image acquisition times, minimizes expensive hardware requirements and reduces motion artifacts in OCT images.
Towards Label-Free 3D Segmentation of Optical Coherence Tomography Images of the Optic Nerve Head Using Deep Learning
Feb 22, 2020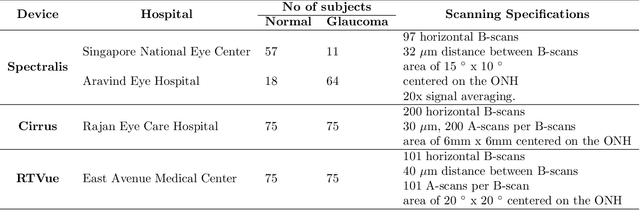
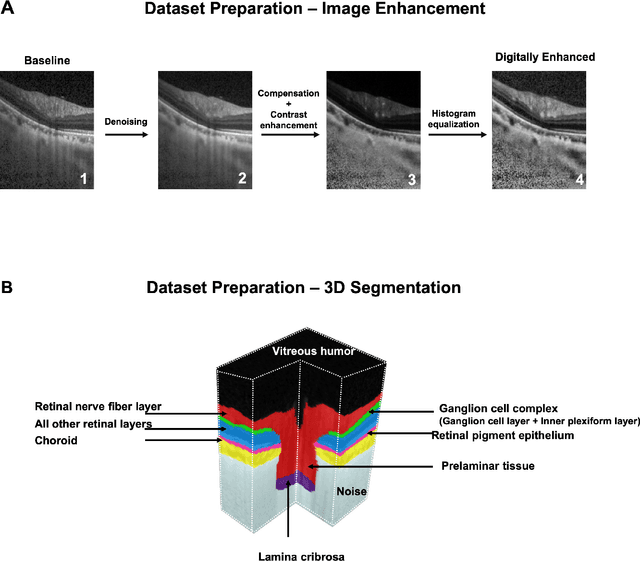
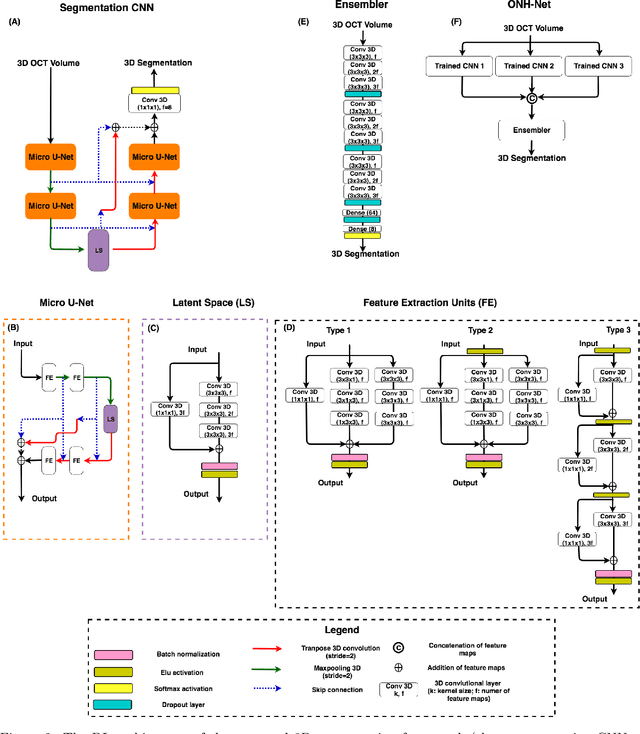
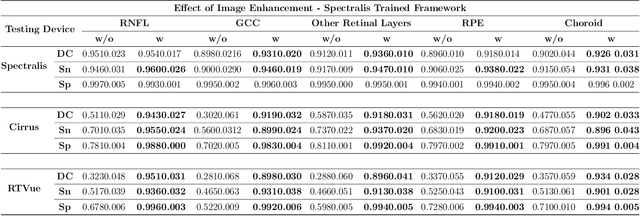
Abstract:Since the introduction of optical coherence tomography (OCT), it has been possible to study the complex 3D morphological changes of the optic nerve head (ONH) tissues that occur along with the progression of glaucoma. Although several deep learning (DL) techniques have been recently proposed for the automated extraction (segmentation) and quantification of these morphological changes, the device specific nature and the difficulty in preparing manual segmentations (training data) limit their clinical adoption. With several new manufacturers and next-generation OCT devices entering the market, the complexity in deploying DL algorithms clinically is only increasing. To address this, we propose a DL based 3D segmentation framework that is easily translatable across OCT devices in a label-free manner (i.e. without the need to manually re-segment data for each device). Specifically, we developed 2 sets of DL networks. The first (referred to as the enhancer) was able to enhance OCT image quality from 3 OCT devices, and harmonized image-characteristics across these devices. The second performed 3D segmentation of 6 important ONH tissue layers. We found that the use of the enhancer was critical for our segmentation network to achieve device independency. In other words, our 3D segmentation network trained on any of 3 devices successfully segmented ONH tissue layers from the other two devices with high performance (Dice coefficients > 0.92). With such an approach, we could automatically segment images from new OCT devices without ever needing manual segmentation data from such devices.
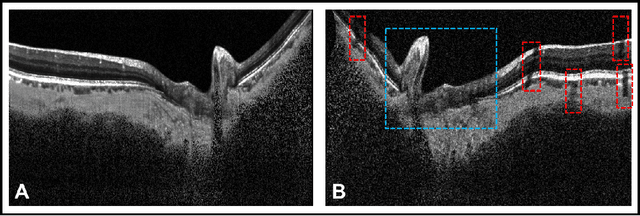
DeshadowGAN: A Deep Learning Approach to Remove Shadows from Optical Coherence Tomography Images
Oct 07, 2019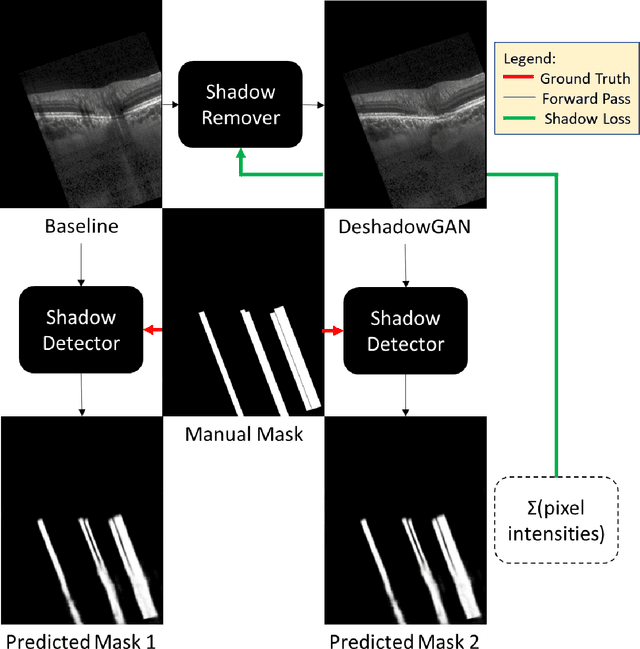
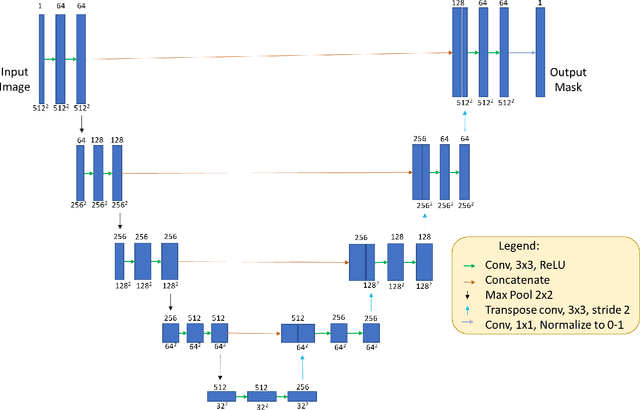
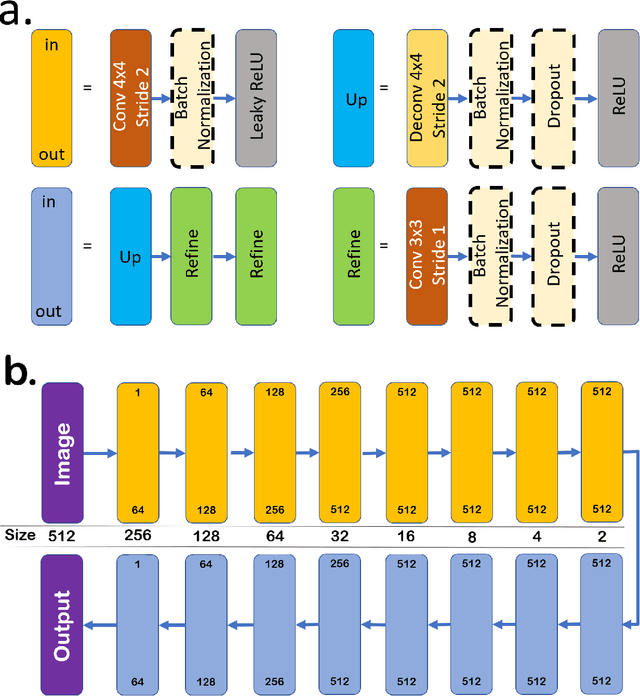
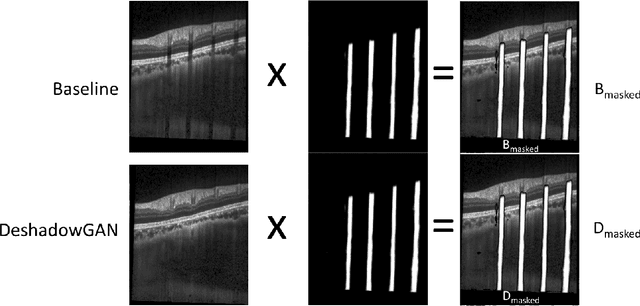
Abstract:Purpose: To remove retinal shadows from optical coherence tomography (OCT) images of the optic nerve head(ONH). Methods:2328 OCT images acquired through the center of the ONH using a Spectralis OCT machine for both eyes of 13 subjects were used to train a generative adversarial network (GAN) using a custom loss function. Image quality was assessed qualitatively (for artifacts) and quantitatively using the intralayer contrast: a measure of shadow visibility ranging from 0 (shadow-free) to 1 (strong shadow) and compared to compensated images. This was computed in the Retinal Nerve Fiber Layer (RNFL), the Inner Plexiform Layer (IPL), the Photoreceptor layer (PR) and the Retinal Pigment Epithelium (RPE) layers. Results: Output images had improved intralayer contrast in all ONH tissue layers. On average the intralayer contrast decreased by 33.7$\pm$6.81%, 28.8$\pm$10.4%, 35.9$\pm$13.0%, and43.0$\pm$19.5%for the RNFL, IPL, PR, and RPE layers respectively, indicating successful shadow removal across all depths. This compared to 70.3$\pm$22.7%, 33.9$\pm$11.5%, 47.0$\pm$11.2%, 26.7$\pm$19.0%for compensation. Output images were also free from artifacts commonly observed with compensation. Conclusions: DeshadowGAN significantly corrected blood vessel shadows in OCT images of the ONH. Our algorithm may be considered as a pre-processing step to improve the performance of a wide range of algorithms including those currently being used for OCT image segmentation, denoising, and classification. Translational Relevance: DeshadowGAN could be integrated to existing OCT devices to improve the diagnosis and prognosis of ocular pathologies.
Deep Learning Algorithms to Isolate and Quantify the Structures of the Anterior Segment in Optical Coherence Tomography Images
Sep 01, 2019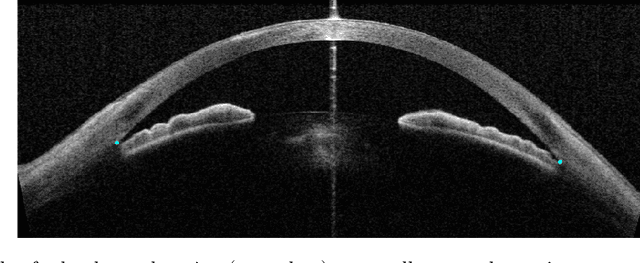
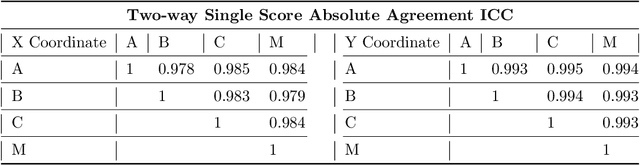
Abstract:Accurate isolation and quantification of intraocular dimensions in the anterior segment (AS) of the eye using optical coherence tomography (OCT) images is important in the diagnosis and treatment of many eye diseases, especially angle closure glaucoma. In this study, we developed a deep convolutional neural network (DCNN) for the localization of the scleral spur, and the segmentation of anterior segment structures (iris, corneo-sclera shell, anterior chamber). With limited training data, the DCNN was able to detect the scleral spur on unseen ASOCT images as accurately as an experienced ophthalmologist; and simultaneously isolated the anterior segment structures with a Dice coefficient of 95.7%. We then automatically extracted eight clinically relevant ASOCT parameters and proposed an automated quality check process that asserts the reliability of these parameters. When combined with an OCT machine capable of imaging multiple radial sections, the algorithms can provide a more complete objective assessment. This is an essential step toward providing a robust automated framework for reliable quantification of ASOCT scans, for applications in the diagnosis and management of angle closure glaucoma.
A Deep Learning Approach to Denoise Optical Coherence Tomography Images of the Optic Nerve Head
Sep 27, 2018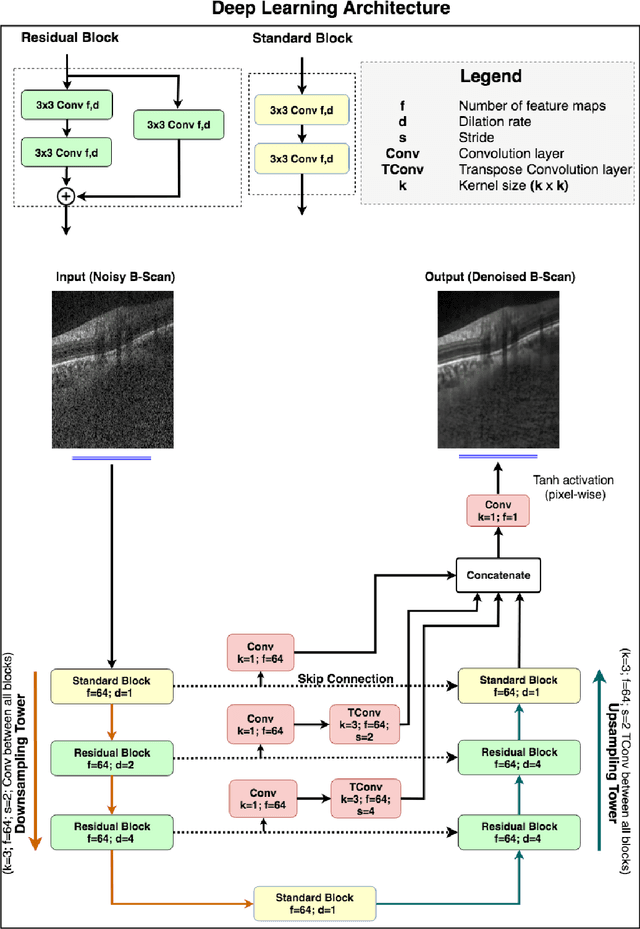
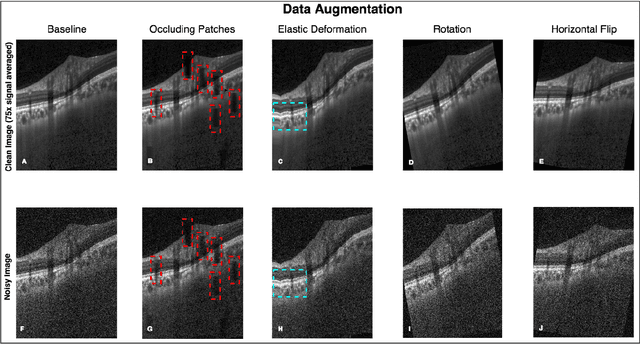
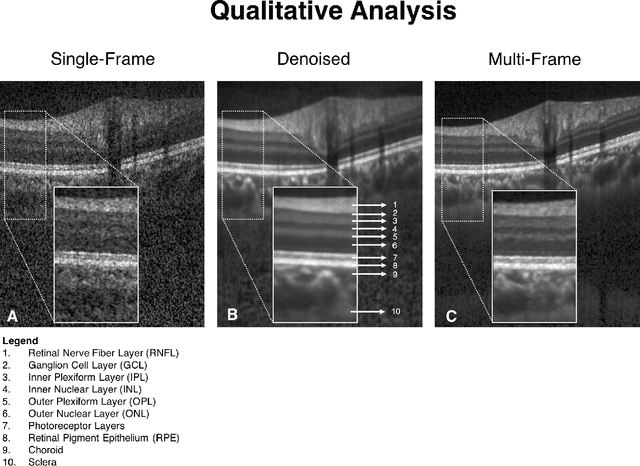
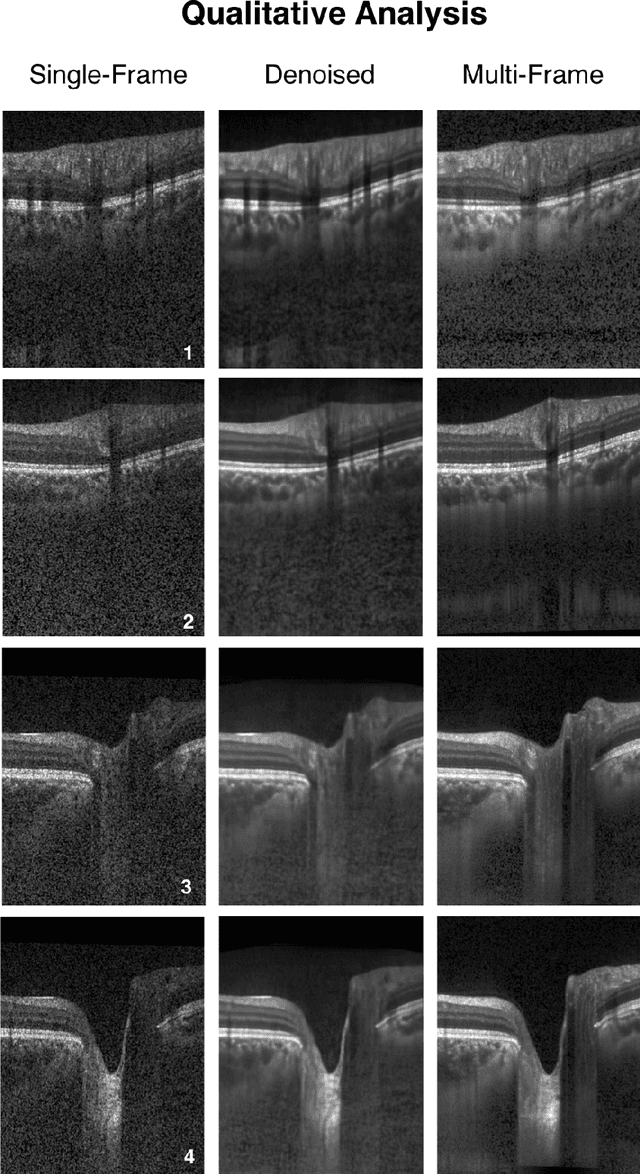
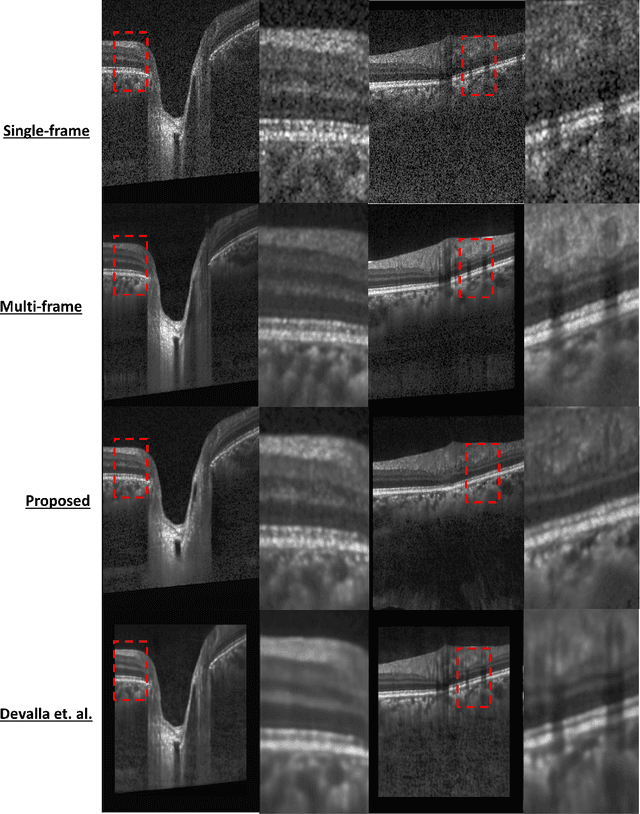
Abstract:Purpose: To develop a deep learning approach to de-noise optical coherence tomography (OCT) B-scans of the optic nerve head (ONH). Methods: Volume scans consisting of 97 horizontal B-scans were acquired through the center of the ONH using a commercial OCT device (Spectralis) for both eyes of 20 subjects. For each eye, single-frame (without signal averaging), and multi-frame (75x signal averaging) volume scans were obtained. A custom deep learning network was then designed and trained with 2,328 "clean B-scans" (multi-frame B-scans), and their corresponding "noisy B-scans" (clean B-scans + gaussian noise) to de-noise the single-frame B-scans. The performance of the de-noising algorithm was assessed qualitatively, and quantitatively on 1,552 B-scans using the signal to noise ratio (SNR), contrast to noise ratio (CNR), and mean structural similarity index metrics (MSSIM). Results: The proposed algorithm successfully denoised unseen single-frame OCT B-scans. The denoised B-scans were qualitatively similar to their corresponding multi-frame B-scans, with enhanced visibility of the ONH tissues. The mean SNR increased from $4.02 \pm 0.68$ dB (single-frame) to $8.14 \pm 1.03$ dB (denoised). For all the ONH tissues, the mean CNR increased from $3.50 \pm 0.56$ (single-frame) to $7.63 \pm 1.81$ (denoised). The MSSIM increased from $0.13 \pm 0.02$ (single frame) to $0.65 \pm 0.03$ (denoised) when compared with the corresponding multi-frame B-scans. Conclusions: Our deep learning algorithm can denoise a single-frame OCT B-scan of the ONH in under 20 ms, thus offering a framework to obtain superior quality OCT B-scans with reduced scanning times and minimal patient discomfort.
DRUNET: A Dilated-Residual U-Net Deep Learning Network to Digitally Stain Optic Nerve Head Tissues in Optical Coherence Tomography Images
Mar 01, 2018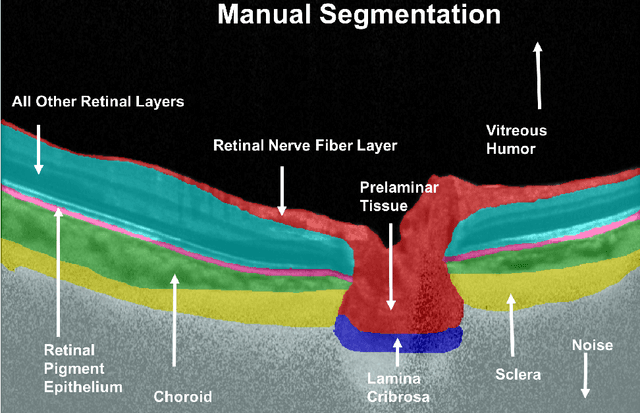
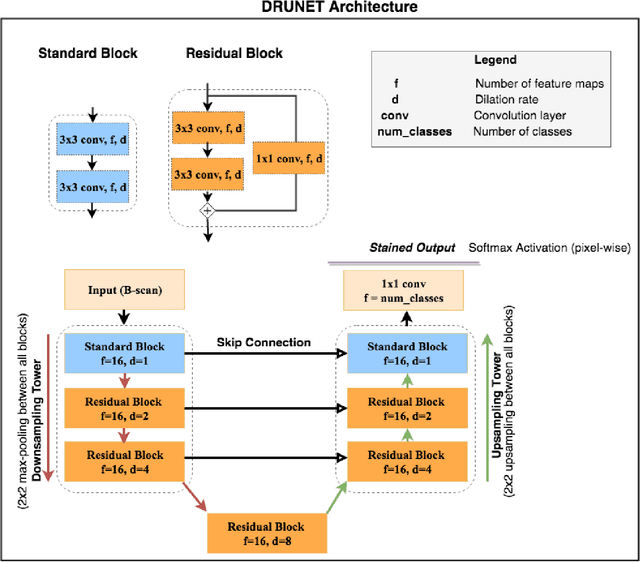
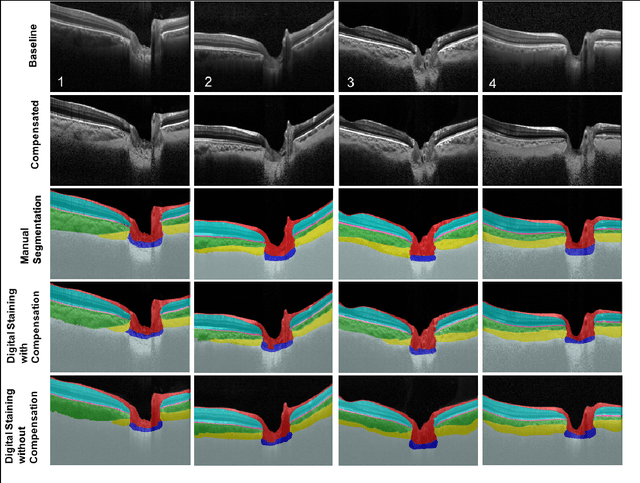
Abstract:Given that the neural and connective tissues of the optic nerve head (ONH) exhibit complex morphological changes with the development and progression of glaucoma, their simultaneous isolation from optical coherence tomography (OCT) images may be of great interest for the clinical diagnosis and management of this pathology. A deep learning algorithm was designed and trained to digitally stain (i.e. highlight) 6 ONH tissue layers by capturing both the local (tissue texture) and contextual information (spatial arrangement of tissues). The overall dice coefficient (mean of all tissues) was $0.91 \pm 0.05$ when assessed against manual segmentations performed by an expert observer. We offer here a robust segmentation framework that could be extended for the automated parametric study of the ONH tissues.
A Deep Learning Approach to Digitally Stain Optical Coherence Tomography Images of the Optic Nerve Head
Jul 24, 2017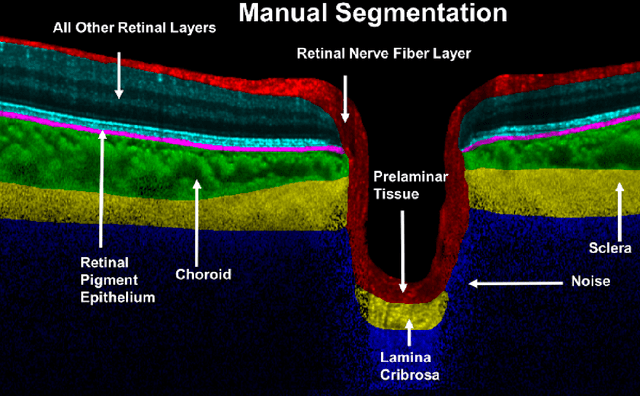
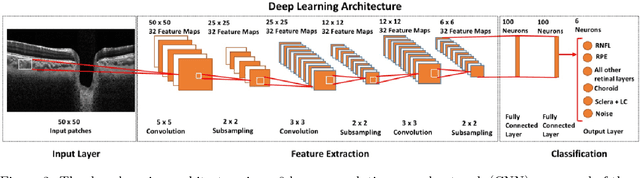
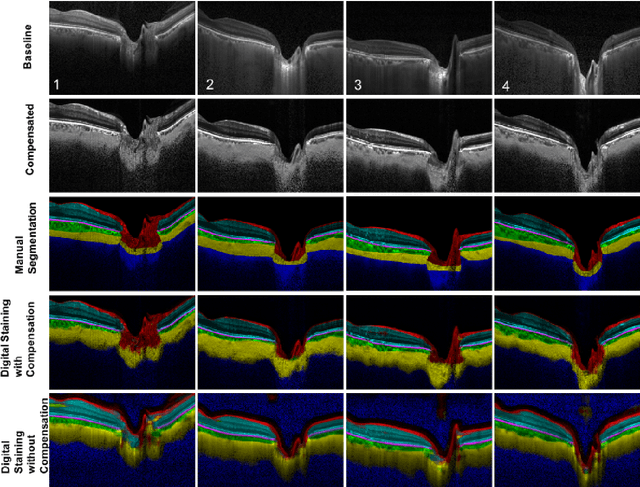
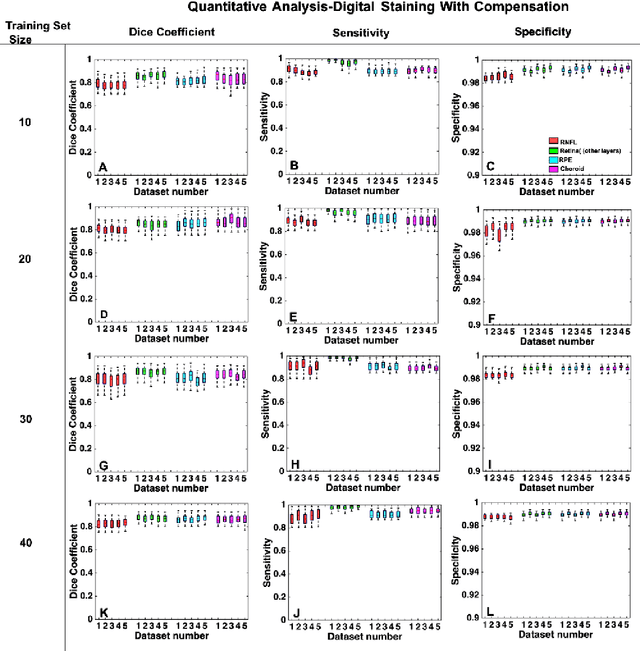
Abstract:Purpose: To develop a deep learning approach to digitally-stain optical coherence tomography (OCT) images of the optic nerve head (ONH). Methods: A horizontal B-scan was acquired through the center of the ONH using OCT (Spectralis) for 1 eye of each of 100 subjects (40 normal & 60 glaucoma). All images were enhanced using adaptive compensation. A custom deep learning network was then designed and trained with the compensated images to digitally stain (i.e. highlight) 6 tissue layers of the ONH. The accuracy of our algorithm was assessed (against manual segmentations) using the Dice coefficient, sensitivity, and specificity. We further studied how compensation and the number of training images affected the performance of our algorithm. Results: For images it had not yet assessed, our algorithm was able to digitally stain the retinal nerve fiber layer + prelamina, the retinal pigment epithelium, all other retinal layers, the choroid, and the peripapillary sclera and lamina cribrosa. For all tissues, the mean dice coefficient was $0.84 \pm 0.03$, the mean sensitivity $0.92 \pm 0.03$, and the mean specificity $0.99 \pm 0.00$. Our algorithm performed significantly better when compensated images were used for training. Increasing the number of images (from 10 to 40) to train our algorithm did not significantly improve performance, except for the RPE. Conclusion. Our deep learning algorithm can simultaneously stain neural and connective tissues in ONH images. Our approach offers a framework to automatically measure multiple key structural parameters of the ONH that may be critical to improve glaucoma management.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge