Giridhar Subramanian
Towards Label-Free 3D Segmentation of Optical Coherence Tomography Images of the Optic Nerve Head Using Deep Learning
Feb 22, 2020
Abstract:Since the introduction of optical coherence tomography (OCT), it has been possible to study the complex 3D morphological changes of the optic nerve head (ONH) tissues that occur along with the progression of glaucoma. Although several deep learning (DL) techniques have been recently proposed for the automated extraction (segmentation) and quantification of these morphological changes, the device specific nature and the difficulty in preparing manual segmentations (training data) limit their clinical adoption. With several new manufacturers and next-generation OCT devices entering the market, the complexity in deploying DL algorithms clinically is only increasing. To address this, we propose a DL based 3D segmentation framework that is easily translatable across OCT devices in a label-free manner (i.e. without the need to manually re-segment data for each device). Specifically, we developed 2 sets of DL networks. The first (referred to as the enhancer) was able to enhance OCT image quality from 3 OCT devices, and harmonized image-characteristics across these devices. The second performed 3D segmentation of 6 important ONH tissue layers. We found that the use of the enhancer was critical for our segmentation network to achieve device independency. In other words, our 3D segmentation network trained on any of 3 devices successfully segmented ONH tissue layers from the other two devices with high performance (Dice coefficients > 0.92). With such an approach, we could automatically segment images from new OCT devices without ever needing manual segmentation data from such devices.
A Deep Learning Approach to Denoise Optical Coherence Tomography Images of the Optic Nerve Head
Sep 27, 2018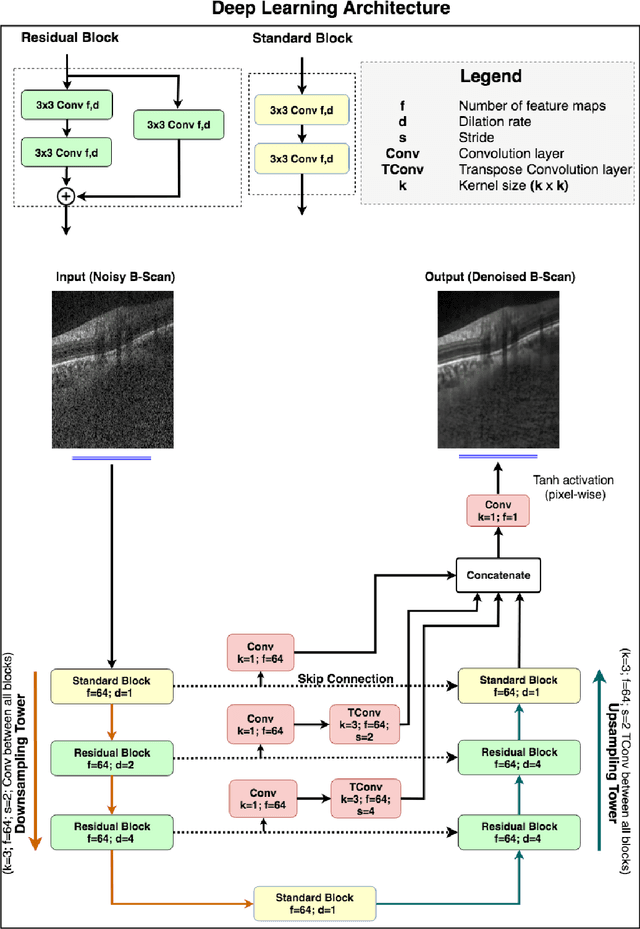
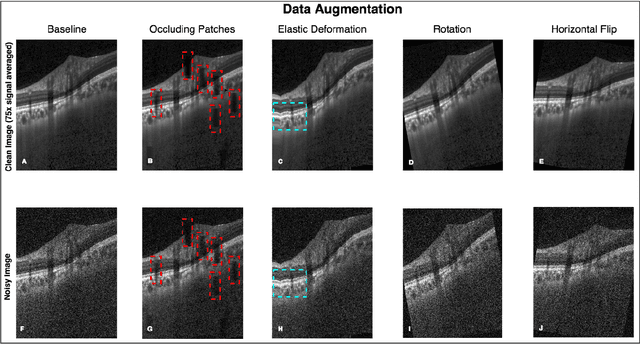
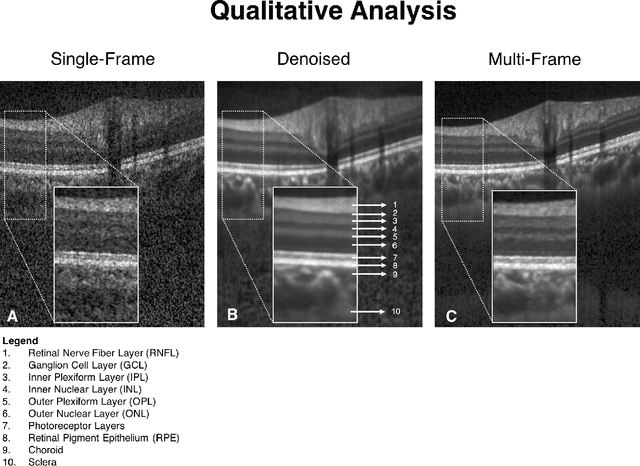
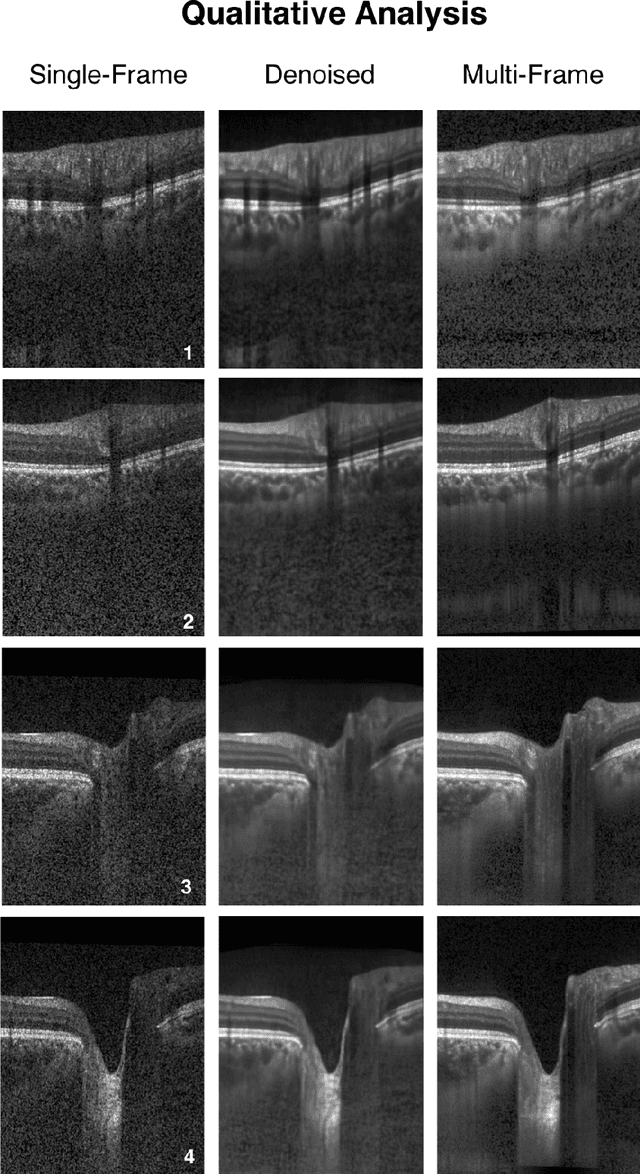
Abstract:Purpose: To develop a deep learning approach to de-noise optical coherence tomography (OCT) B-scans of the optic nerve head (ONH). Methods: Volume scans consisting of 97 horizontal B-scans were acquired through the center of the ONH using a commercial OCT device (Spectralis) for both eyes of 20 subjects. For each eye, single-frame (without signal averaging), and multi-frame (75x signal averaging) volume scans were obtained. A custom deep learning network was then designed and trained with 2,328 "clean B-scans" (multi-frame B-scans), and their corresponding "noisy B-scans" (clean B-scans + gaussian noise) to de-noise the single-frame B-scans. The performance of the de-noising algorithm was assessed qualitatively, and quantitatively on 1,552 B-scans using the signal to noise ratio (SNR), contrast to noise ratio (CNR), and mean structural similarity index metrics (MSSIM). Results: The proposed algorithm successfully denoised unseen single-frame OCT B-scans. The denoised B-scans were qualitatively similar to their corresponding multi-frame B-scans, with enhanced visibility of the ONH tissues. The mean SNR increased from $4.02 \pm 0.68$ dB (single-frame) to $8.14 \pm 1.03$ dB (denoised). For all the ONH tissues, the mean CNR increased from $3.50 \pm 0.56$ (single-frame) to $7.63 \pm 1.81$ (denoised). The MSSIM increased from $0.13 \pm 0.02$ (single frame) to $0.65 \pm 0.03$ (denoised) when compared with the corresponding multi-frame B-scans. Conclusions: Our deep learning algorithm can denoise a single-frame OCT B-scan of the ONH in under 20 ms, thus offering a framework to obtain superior quality OCT B-scans with reduced scanning times and minimal patient discomfort.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge