Alexandre H. Thiery
Out-of-Distribution Detection with a Single Unconditional Diffusion Model
May 20, 2024



Abstract:Out-of-distribution (OOD) detection is a critical task in machine learning that seeks to identify abnormal samples. Traditionally, unsupervised methods utilize a deep generative model for OOD detection. However, such approaches necessitate a different model when evaluating abnormality against a new distribution. With the emergence of foundational generative models, this paper explores whether a single generalist model can also perform OOD detection across diverse tasks. To that end, we introduce our method, Diffusion Paths, (DiffPath) in this work. DiffPath proposes to utilize a single diffusion model originally trained to perform unconditional generation for OOD detection. Specifically, we introduce a novel technique of measuring the rate-of-change and curvature of the diffusion paths connecting samples to the standard normal. Extensive experiments show that with a single model, DiffPath outperforms prior work on a variety of OOD tasks involving different distributions. Our code is publicly available at https://github.com/clear-nus/diffpath.
AI-based Clinical Assessment of Optic Nerve Head Robustness Superseding Biomechanical Testing
Jun 09, 2022
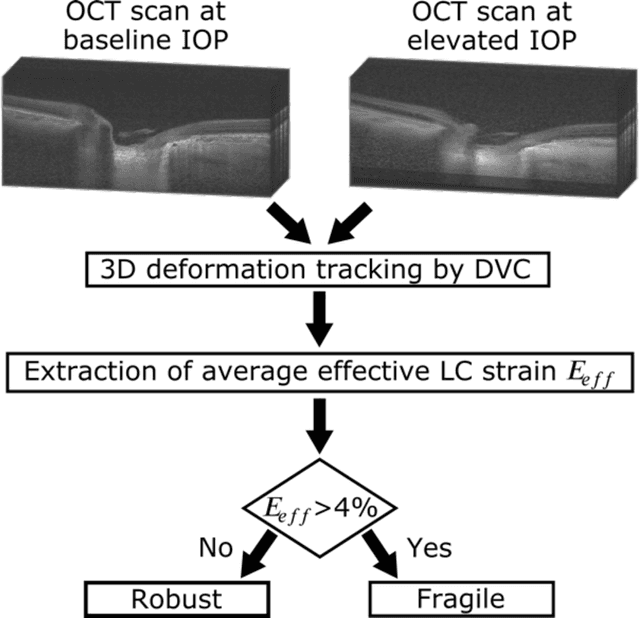
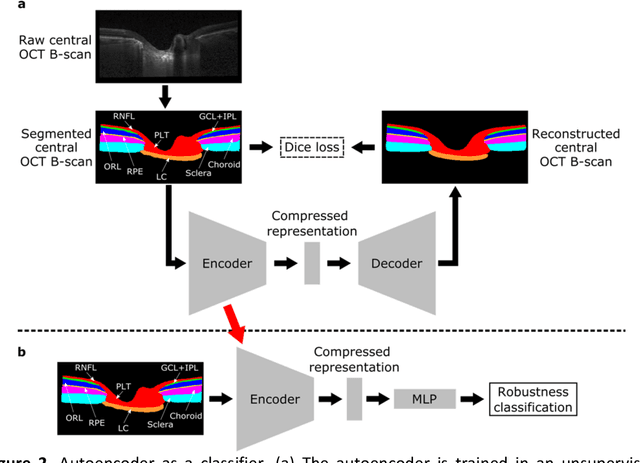
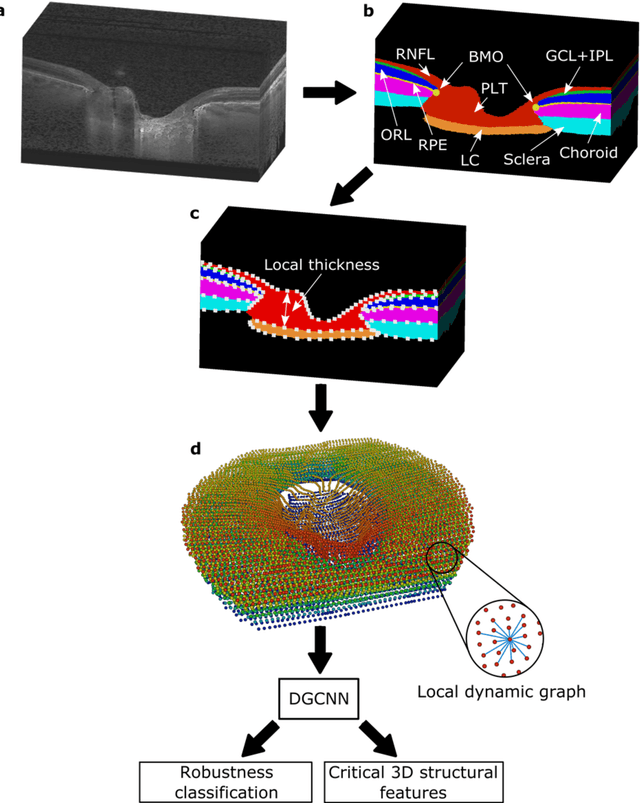
Abstract:$\mathbf{Purpose}$: To use artificial intelligence (AI) to: (1) exploit biomechanical knowledge of the optic nerve head (ONH) from a relatively large population; (2) assess ONH robustness from a single optical coherence tomography (OCT) scan of the ONH; (3) identify what critical three-dimensional (3D) structural features make a given ONH robust. $\mathbf{Design}$: Retrospective cross-sectional study. $\mathbf{Methods}$: 316 subjects had their ONHs imaged with OCT before and after acute intraocular pressure (IOP) elevation through ophthalmo-dynamometry. IOP-induced lamina-cribrosa deformations were then mapped in 3D and used to classify ONHs. Those with LC deformations superior to 4% were considered fragile, while those with deformations inferior to 4% robust. Learning from these data, we compared three AI algorithms to predict ONH robustness strictly from a baseline (undeformed) OCT volume: (1) a random forest classifier; (2) an autoencoder; and (3) a dynamic graph CNN (DGCNN). The latter algorithm also allowed us to identify what critical 3D structural features make a given ONH robust. $\mathbf{Results}$: All 3 methods were able to predict ONH robustness from 3D structural information alone and without the need to perform biomechanical testing. The DGCNN (area under the receiver operating curve [AUC]: 0.76 $\pm$ 0.08) outperformed the autoencoder (AUC: 0.70 $\pm$ 0.07) and the random forest classifier (AUC: 0.69 $\pm$ 0.05). Interestingly, to assess ONH robustness, the DGCNN mainly used information from the scleral canal and the LC insertion sites. $\mathbf{Conclusions}$: We propose an AI-driven approach that can assess the robustness of a given ONH solely from a single OCT scan of the ONH, and without the need to perform biomechanical testing. Longitudinal studies should establish whether ONH robustness could help us identify fast visual field loss progressors.
Computational Doob's $h$-transforms for Online Filtering of Discretely Observed Diffusions
Jun 07, 2022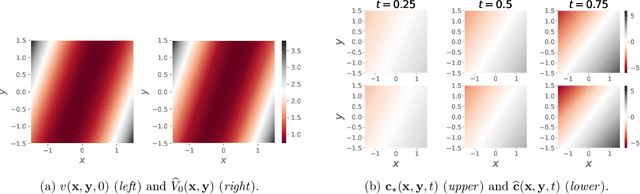


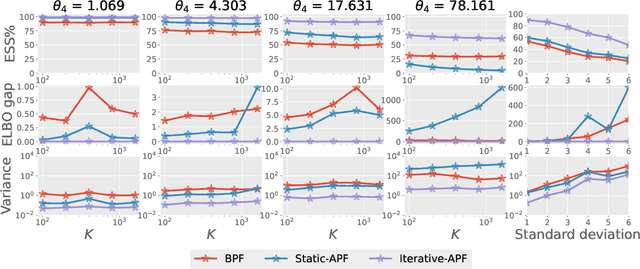
Abstract:This paper is concerned with online filtering of discretely observed nonlinear diffusion processes. Our approach is based on the fully adapted auxiliary particle filter, which involves Doob's $h$-transforms that are typically intractable. We propose a computational framework to approximate these $h$-transforms by solving the underlying backward Kolmogorov equations using nonlinear Feynman-Kac formulas and neural networks. The methodology allows one to train a locally optimal particle filter prior to the data-assimilation procedure. Numerical experiments illustrate that the proposed approach can be orders of magnitude more efficient than the bootstrap particle filter in the regime of highly informative observations, when the observations are extreme under the model, and if the state dimension is large.
Medical Application of Geometric Deep Learning for the Diagnosis of Glaucoma
Apr 14, 2022
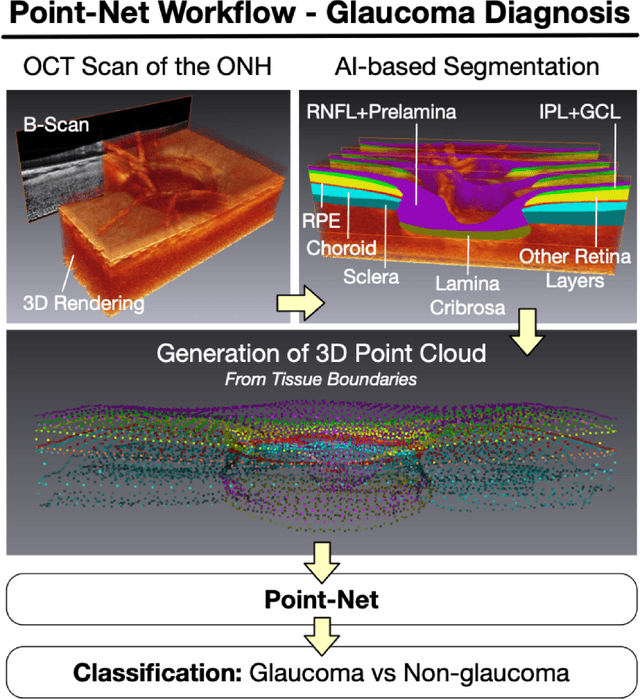
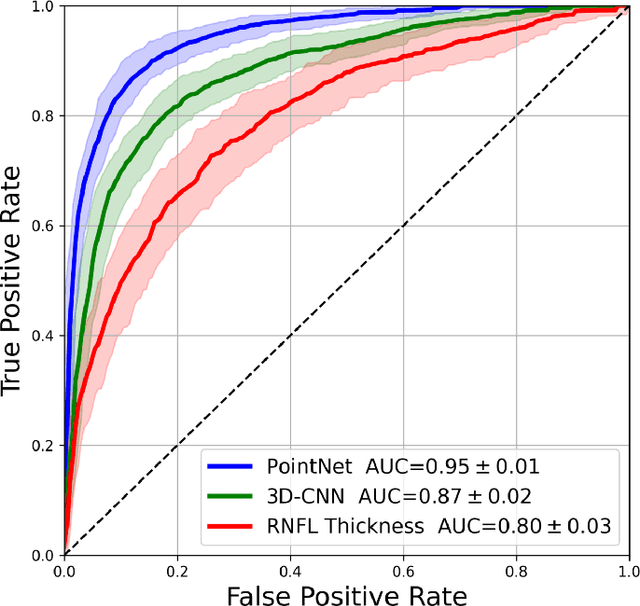
Abstract:Purpose: (1) To assess the performance of geometric deep learning (PointNet) in diagnosing glaucoma from a single optical coherence tomography (OCT) 3D scan of the optic nerve head (ONH); (2) To compare its performance to that obtained with a standard 3D convolutional neural network (CNN), and with a gold-standard glaucoma parameter, i.e. retinal nerve fiber layer (RNFL) thickness. Methods: 3D raster scans of the ONH were acquired with Spectralis OCT for 477 glaucoma and 2,296 non-glaucoma subjects at the Singapore National Eye Centre. All volumes were automatically segmented using deep learning to identify 7 major neural and connective tissues including the RNFL, the prelamina, and the lamina cribrosa (LC). Each ONH was then represented as a 3D point cloud with 1,000 points chosen randomly from all tissue boundaries. To simplify the problem, all ONH point clouds were aligned with respect to the plane and center of Bruch's membrane opening. Geometric deep learning (PointNet) was then used to provide a glaucoma diagnosis from a single OCT point cloud. The performance of our approach was compared to that obtained with a 3D CNN, and with RNFL thickness. Results: PointNet was able to provide a robust glaucoma diagnosis solely from the ONH represented as a 3D point cloud (AUC=95%). The performance of PointNet was superior to that obtained with a standard 3D CNN (AUC=87%) and with that obtained from RNFL thickness alone (AUC=80%). Discussion: We provide a proof-of-principle for the application of geometric deep learning in the field of glaucoma. Our technique requires significantly less information as input to perform better than a 3D CNN, and with an AUC superior to that obtained from RNFL thickness alone. Geometric deep learning may have wide applicability in the field of Ophthalmology.
Explainable and Interpretable Diabetic Retinopathy Classification Based on Neural-Symbolic Learning
Apr 01, 2022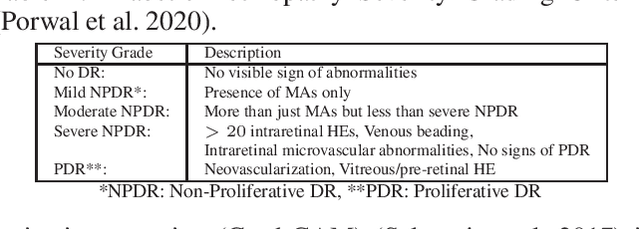
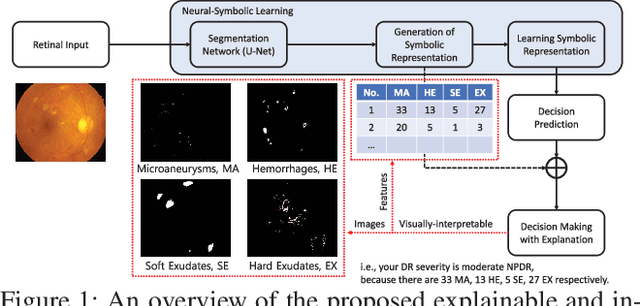
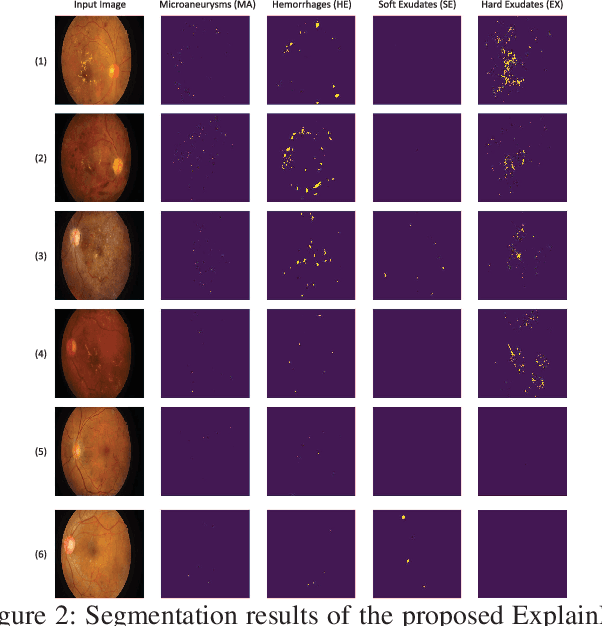

Abstract:In this paper, we propose an explainable and interpretable diabetic retinopathy (ExplainDR) classification model based on neural-symbolic learning. To gain explainability, a highlevel symbolic representation should be considered in decision making. Specifically, we introduce a human-readable symbolic representation, which follows a taxonomy style of diabetic retinopathy characteristics related to eye health conditions to achieve explainability. We then include humanreadable features obtained from the symbolic representation in the disease prediction. Experimental results on a diabetic retinopathy classification dataset show that our proposed ExplainDR method exhibits promising performance when compared to that from state-of-the-art methods applied to the IDRiD dataset, while also providing interpretability and explainability.
The Three-Dimensional Structural Configuration of the Central Retinal Vessel Trunk and Branches as a Glaucoma Biomarker
Nov 09, 2021
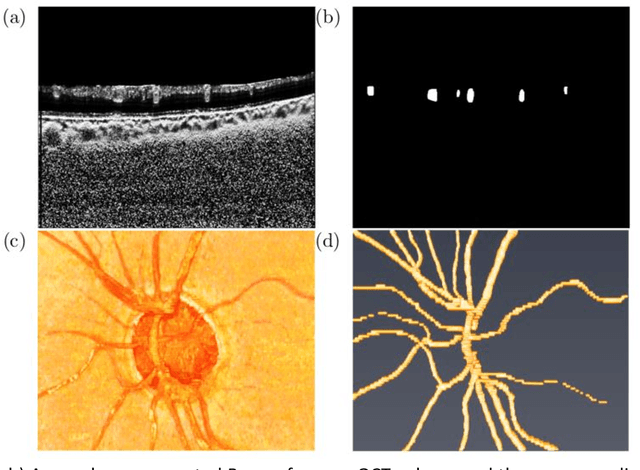
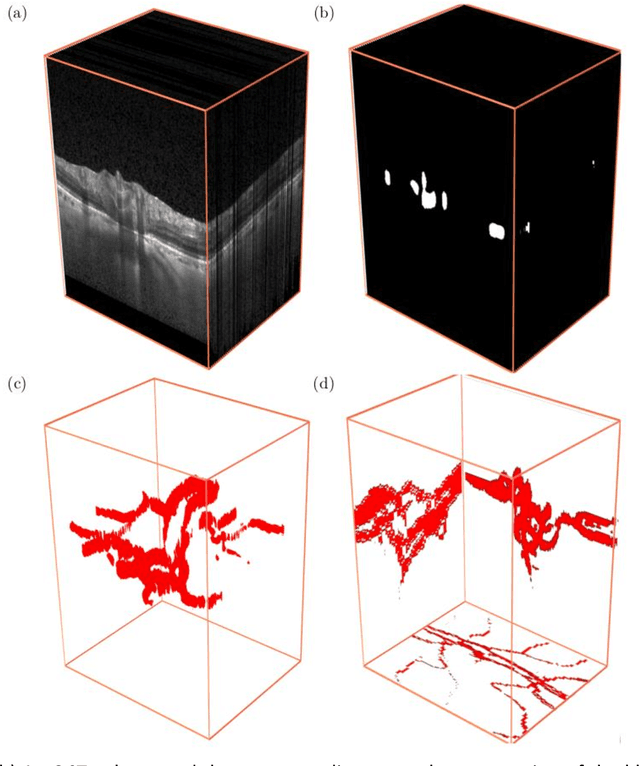

Abstract:Purpose: To assess whether the three-dimensional (3D) structural configuration of the central retinal vessel trunk and its branches (CRVT&B) could be used as a diagnostic marker for glaucoma. Method: We trained a deep learning network to automatically segment the CRVT&B from the B-scans of the optical coherence tomography (OCT) volume of the optic nerve head (ONH). Subsequently, two different approaches were used for glaucoma diagnosis using the structural configuration of the CRVT&B as extracted from the OCT volumes. In the first approach, we aimed to provide a diagnosis using only 3D CNN and the 3D structure of the CRVT&B. For the second approach, we projected the 3D structure of the CRVT&B orthographically onto three planes to obtain 2D images, and then a 2D CNN was used for diagnosis. The segmentation accuracy was evaluated using the Dice coefficient, whereas the diagnostic accuracy was assessed using the area under the receiver operating characteristic curves (AUC). The diagnostic performance of the CRVT&B was also compared with that of retinal nerve fiber layer (RNFL) thickness. Results: Our segmentation network was able to efficiently segment retinal blood vessels from OCT scans. On a test set, we achieved a Dice coefficient of 0.81\pm0.07. The 3D and 2D diagnostic networks were able to differentiate glaucoma from non-glaucoma subjects with accuracies of 82.7% and 83.3%, respectively. The corresponding AUCs for CRVT&B were 0.89 and 0.90, higher than those obtained with RNFL thickness alone. Conclusions: Our work demonstrated that the diagnostic power of the CRVT&B is superior to that of a gold-standard glaucoma parameter, i.e., RNFL thickness. Our work also suggested that the major retinal blood vessels form a skeleton -- the configuration of which may be representative of major ONH structural changes as typically observed with the development and progression of glaucoma.
Pretrained equivariant features improve unsupervised landmark discovery
Apr 07, 2021
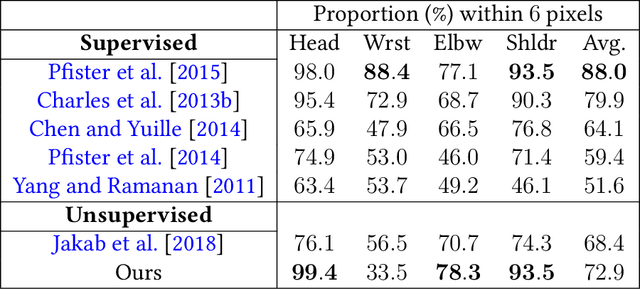
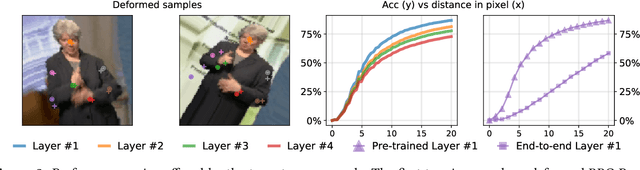

Abstract:Locating semantically meaningful landmark points is a crucial component of a large number of computer vision pipelines. Because of the small number of available datasets with ground truth landmark annotations, it is important to design robust unsupervised and semi-supervised methods for landmark detection. Many of the recent unsupervised learning methods rely on the equivariance properties of landmarks to synthetic image deformations. Our work focuses on such widely used methods and sheds light on its core problem, its inability to produce equivariant intermediate convolutional features. This finding leads us to formulate a two-step unsupervised approach that overcomes this challenge by first learning powerful pixel-based features and then use the pre-trained features to learn a landmark detector by the traditional equivariance method. Our method produces state-of-the-art results in several challenging landmark detection datasets such as the BBC Pose dataset and the Cat-Head dataset. It performs comparably on a range of other benchmarks.
On Data-Augmentation and Consistency-Based Semi-Supervised Learning
Jan 18, 2021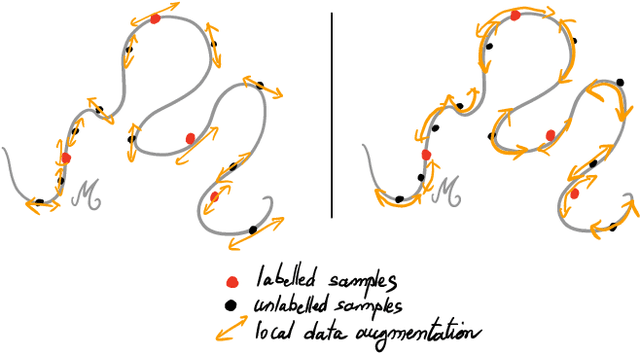
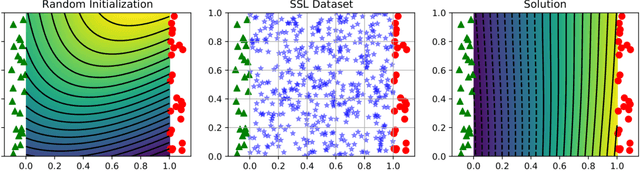
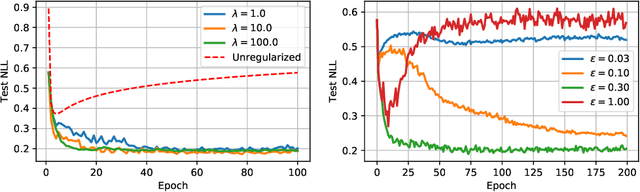
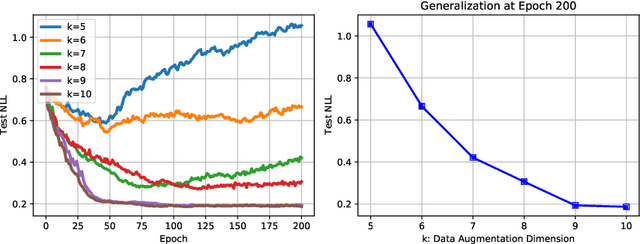
Abstract:Recently proposed consistency-based Semi-Supervised Learning (SSL) methods such as the $\Pi$-model, temporal ensembling, the mean teacher, or the virtual adversarial training, have advanced the state of the art in several SSL tasks. These methods can typically reach performances that are comparable to their fully supervised counterparts while using only a fraction of labelled examples. Despite these methodological advances, the understanding of these methods is still relatively limited. In this text, we analyse (variations of) the $\Pi$-model in settings where analytically tractable results can be obtained. We establish links with Manifold Tangent Classifiers and demonstrate that the quality of the perturbations is key to obtaining reasonable SSL performances. Importantly, we propose a simple extension of the Hidden Manifold Model that naturally incorporates data-augmentation schemes and offers a framework for understanding and experimenting with SSL methods.
Uncertainty Quantification and Deep Ensembles
Jul 20, 2020



Abstract:Deep Learning methods are known to suffer from calibration issues: they typically produce over-confident estimates. These problems are exacerbated in the low data regime. Although the calibration of probabilistic models is well studied, calibrating extremely over-parametrized models in the low-data regime presents unique challenges. We show that deep-ensembles do not necessarily lead to improved calibration properties. In fact, we show that standard ensembling methods, when used in conjunction with modern techniques such as mixup regularization, can lead to less calibrated models. In this text, we examine the interplay between three of the most simple and commonly used approaches to leverage deep learning when data is scarce: data-augmentation, ensembling, and post-processing calibration methods. We demonstrate that, although standard ensembling techniques certainly help to boost accuracy, the calibration of deep-ensembles relies on subtle trade-offs. Our main finding is that calibration methods such as temperature scaling need to be slightly tweaked when used with deep-ensembles and, crucially, need to be executed after the averaging process. Our simulations indicate that, in the low data regime, this simple strategy can halve the Expected Calibration Error (ECE) on a range of benchmark classification problems when compared to standard deep-ensembles.
Towards Label-Free 3D Segmentation of Optical Coherence Tomography Images of the Optic Nerve Head Using Deep Learning
Feb 22, 2020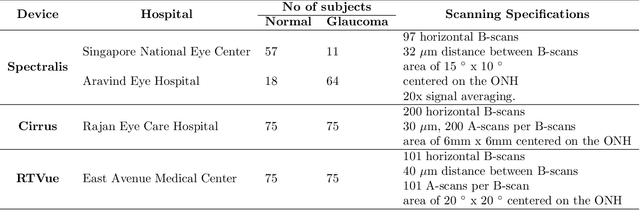
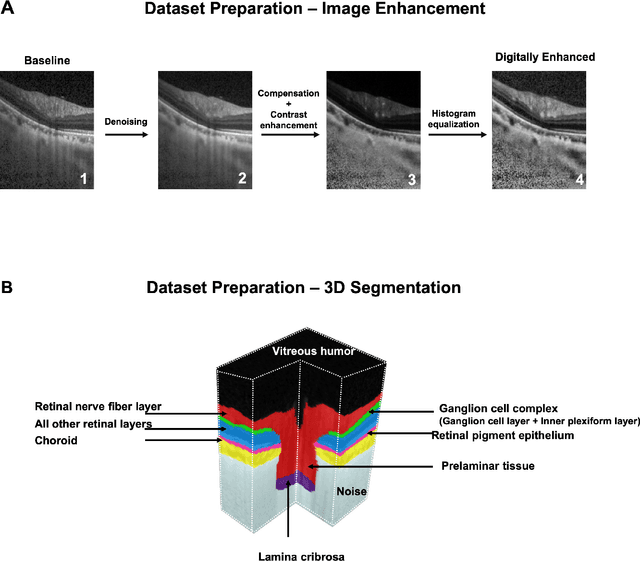
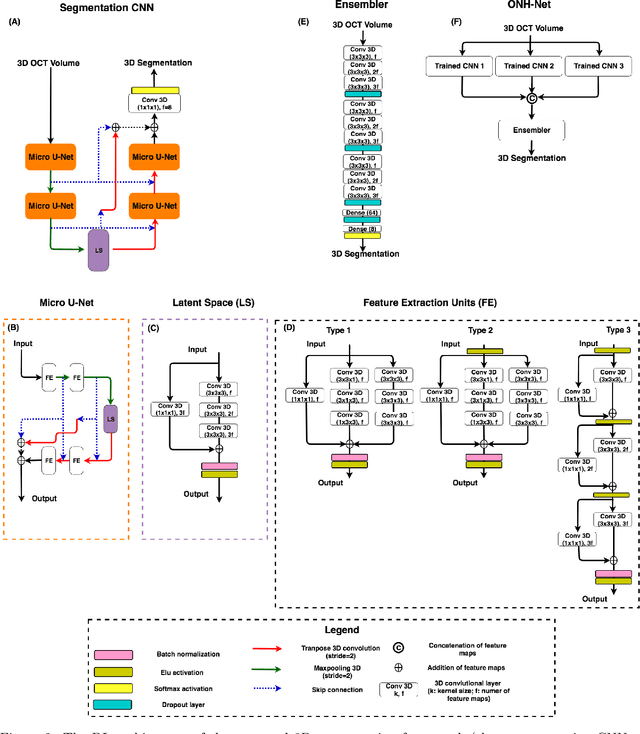
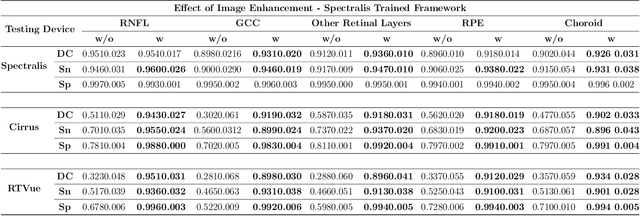
Abstract:Since the introduction of optical coherence tomography (OCT), it has been possible to study the complex 3D morphological changes of the optic nerve head (ONH) tissues that occur along with the progression of glaucoma. Although several deep learning (DL) techniques have been recently proposed for the automated extraction (segmentation) and quantification of these morphological changes, the device specific nature and the difficulty in preparing manual segmentations (training data) limit their clinical adoption. With several new manufacturers and next-generation OCT devices entering the market, the complexity in deploying DL algorithms clinically is only increasing. To address this, we propose a DL based 3D segmentation framework that is easily translatable across OCT devices in a label-free manner (i.e. without the need to manually re-segment data for each device). Specifically, we developed 2 sets of DL networks. The first (referred to as the enhancer) was able to enhance OCT image quality from 3 OCT devices, and harmonized image-characteristics across these devices. The second performed 3D segmentation of 6 important ONH tissue layers. We found that the use of the enhancer was critical for our segmentation network to achieve device independency. In other words, our 3D segmentation network trained on any of 3 devices successfully segmented ONH tissue layers from the other two devices with high performance (Dice coefficients > 0.92). With such an approach, we could automatically segment images from new OCT devices without ever needing manual segmentation data from such devices.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge