Matthias Jung
RoentMod: A Synthetic Chest X-Ray Modification Model to Identify and Correct Image Interpretation Model Shortcuts
Sep 10, 2025Abstract:Chest radiographs (CXRs) are among the most common tests in medicine. Automated image interpretation may reduce radiologists\' workload and expand access to diagnostic expertise. Deep learning multi-task and foundation models have shown strong performance for CXR interpretation but are vulnerable to shortcut learning, where models rely on spurious and off-target correlations rather than clinically relevant features to make decisions. We introduce RoentMod, a counterfactual image editing framework that generates anatomically realistic CXRs with user-specified, synthetic pathology while preserving unrelated anatomical features of the original scan. RoentMod combines an open-source medical image generator (RoentGen) with an image-to-image modification model without requiring retraining. In reader studies with board-certified radiologists and radiology residents, RoentMod-produced images appeared realistic in 93\% of cases, correctly incorporated the specified finding in 89-99\% of cases, and preserved native anatomy comparable to real follow-up CXRs. Using RoentMod, we demonstrate that state-of-the-art multi-task and foundation models frequently exploit off-target pathology as shortcuts, limiting their specificity. Incorporating RoentMod-generated counterfactual images during training mitigated this vulnerability, improving model discrimination across multiple pathologies by 3-19\% AUC in internal validation and by 1-11\% for 5 out of 6 tested pathologies in external testing. These findings establish RoentMod as a broadly applicable tool for probing and correcting shortcut learning in medical AI. By enabling controlled counterfactual interventions, RoentMod enhances the robustness and interpretability of CXR interpretation models and provides a generalizable strategy for improving foundation models in medical imaging.
TotalVibeSegmentator: Full Torso Segmentation for the NAKO and UK Biobank in Volumetric Interpolated Breath-hold Examination Body Images
May 31, 2024



Abstract:Objectives: To present a publicly available torso segmentation network for large epidemiology datasets on volumetric interpolated breath-hold examination (VIBE) images. Materials & Methods: We extracted preliminary segmentations from TotalSegmentator, spine, and body composition networks for VIBE images, then improved them iteratively and retrained a nnUNet network. Using subsets of NAKO (85 subjects) and UK Biobank (16 subjects), we evaluated with Dice-score on a holdout set (12 subjects) and existing organ segmentation approach (1000 subjects), generating 71 semantic segmentation types for VIBE images. We provide an additional network for the vertebra segments 22 individual vertebra types. Results: We achieved an average Dice score of 0.89 +- 0.07 overall 71 segmentation labels. We scored > 0.90 Dice-score on the abdominal organs except for the pancreas with a Dice of 0.70. Conclusion: Our work offers a detailed and refined publicly available full torso segmentation on VIBE images.
TotalSegmentator MRI: Sequence-Independent Segmentation of 59 Anatomical Structures in MR images
May 29, 2024



Abstract:Purpose: To develop an open-source and easy-to-use segmentation model that can automatically and robustly segment most major anatomical structures in MR images independently of the MR sequence. Materials and Methods: In this study we extended the capabilities of TotalSegmentator to MR images. 298 MR scans and 227 CT scans were used to segment 59 anatomical structures (20 organs, 18 bones, 11 muscles, 7 vessels, 3 tissue types) relevant for use cases such as organ volumetry, disease characterization, and surgical planning. The MR and CT images were randomly sampled from routine clinical studies and thus represent a real-world dataset (different ages, pathologies, scanners, body parts, sequences, contrasts, echo times, repetition times, field strengths, slice thicknesses and sites). We trained an nnU-Net segmentation algorithm on this dataset and calculated Dice similarity coefficients (Dice) to evaluate the model's performance. Results: The model showed a Dice score of 0.824 (CI: 0.801, 0.842) on the test set, which included a wide range of clinical data with major pathologies. The model significantly outperformed two other publicly available segmentation models (Dice score, 0.824 versus 0.762; p<0.001 and 0.762 versus 0.542; p<0.001). On the CT image test set of the original TotalSegmentator paper it almost matches the performance of the original TotalSegmentator (Dice score, 0.960 versus 0.970; p<0.001). Conclusion: Our proposed model extends the capabilities of TotalSegmentator to MR images. The annotated dataset (https://zenodo.org/doi/10.5281/zenodo.11367004) and open-source toolkit (https://www.github.com/wasserth/TotalSegmentator) are publicly available.
Deep-CLASS at ISIC Machine Learning Challenge 2018
Jul 24, 2018
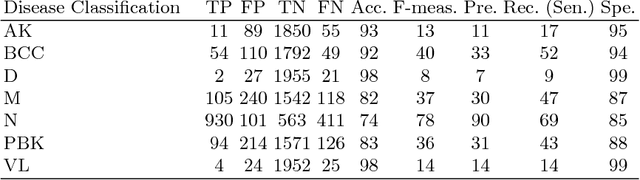
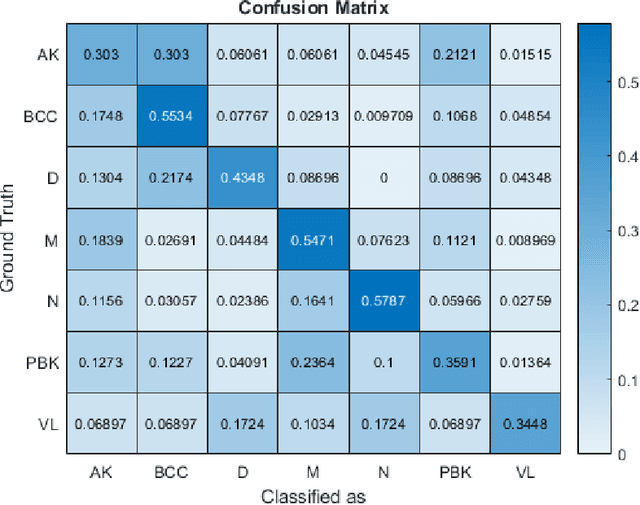
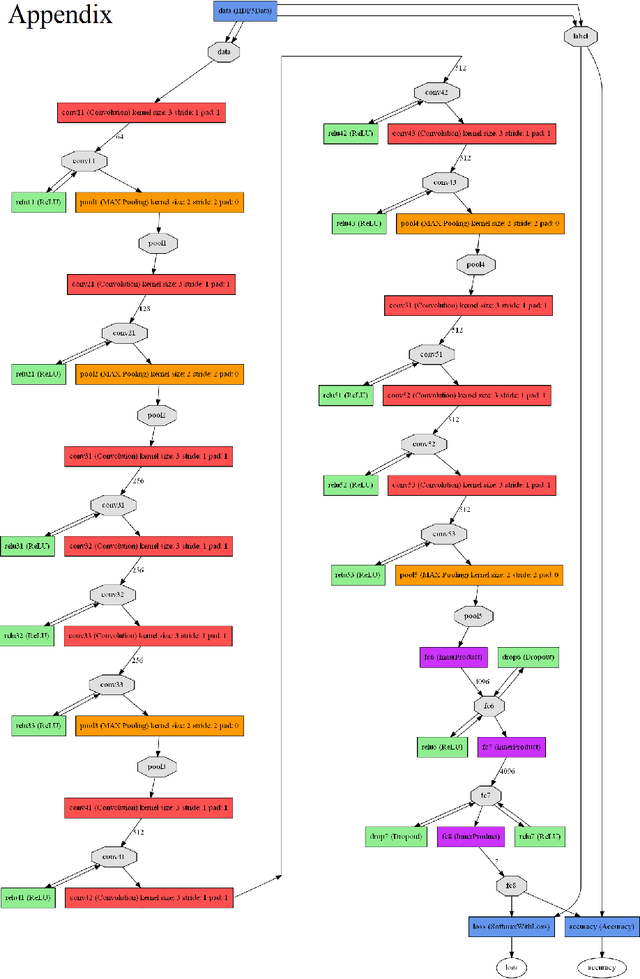
Abstract:This paper reports the method and evaluation results of MedAusbild team for ISIC challenge task. Since early 2017, our team has worked on melanoma classification [1][6], and has employed deep learning since beginning of 2018 [7]. Deep learning helps researchers absolutely to treat and detect diseases by analyzing medical data (e.g., medical images). One of the representative models among the various deep-learning models is a convolutional neural network (CNN). Although our team has an experience with segmentation and classification of benign and malignant skin-lesions, we have participated in the task 3 of ISIC Challenge 2018 for classification of seven skin diseases, explained in this paper.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge