Sophie Starck
CINeMA: Conditional Implicit Neural Multi-Modal Atlas for a Spatio-Temporal Representation of the Perinatal Brain
Jun 11, 2025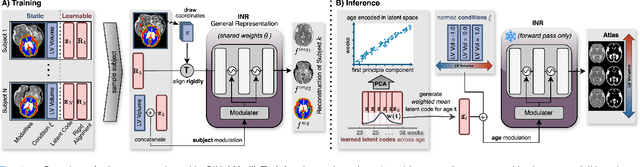
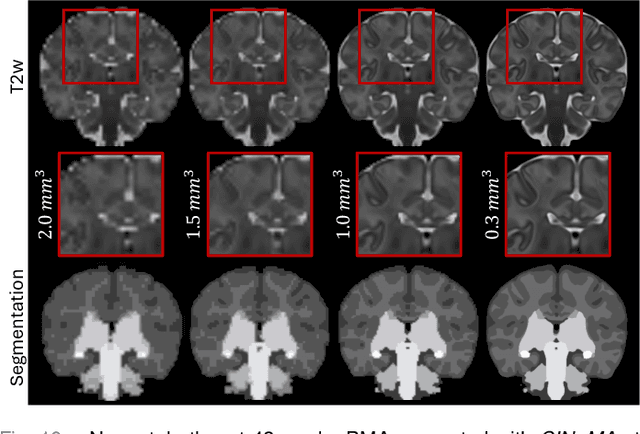
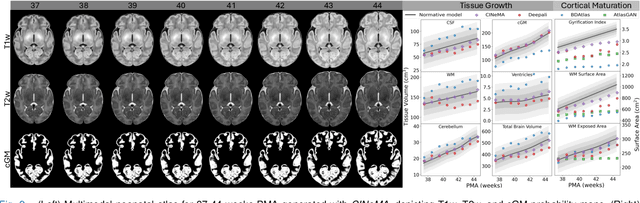
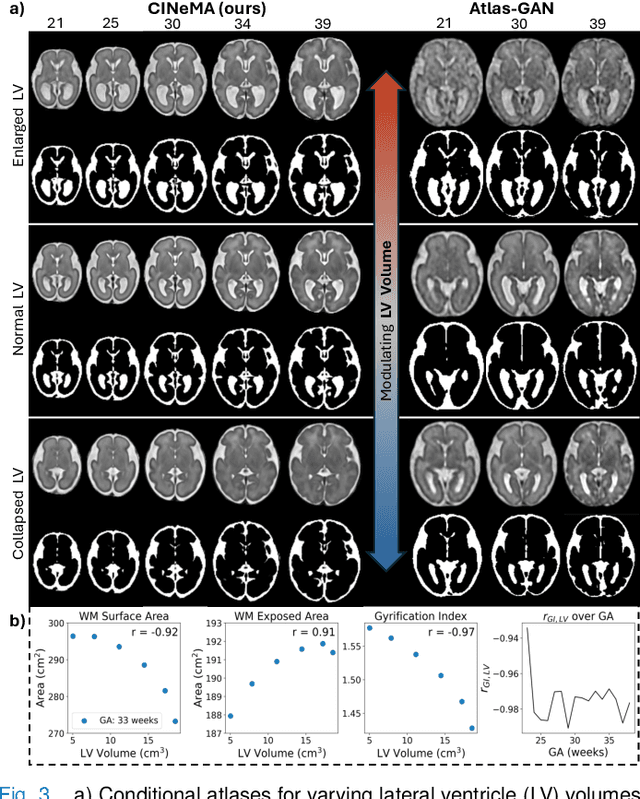
Abstract:Magnetic resonance imaging of fetal and neonatal brains reveals rapid neurodevelopment marked by substantial anatomical changes unfolding within days. Studying this critical stage of the developing human brain, therefore, requires accurate brain models-referred to as atlases-of high spatial and temporal resolution. To meet these demands, established traditional atlases and recently proposed deep learning-based methods rely on large and comprehensive datasets. This poses a major challenge for studying brains in the presence of pathologies for which data remains scarce. We address this limitation with CINeMA (Conditional Implicit Neural Multi-Modal Atlas), a novel framework for creating high-resolution, spatio-temporal, multimodal brain atlases, suitable for low-data settings. Unlike established methods, CINeMA operates in latent space, avoiding compute-intensive image registration and reducing atlas construction times from days to minutes. Furthermore, it enables flexible conditioning on anatomical features including GA, birth age, and pathologies like ventriculomegaly (VM) and agenesis of the corpus callosum (ACC). CINeMA supports downstream tasks such as tissue segmentation and age prediction whereas its generative properties enable synthetic data creation and anatomically informed data augmentation. Surpassing state-of-the-art methods in accuracy, efficiency, and versatility, CINeMA represents a powerful tool for advancing brain research. We release the code and atlases at https://github.com/m-dannecker/CINeMA.
MAGO-SP: Detection and Correction of Water-Fat Swaps in Magnitude-Only VIBE MRI
Feb 20, 2025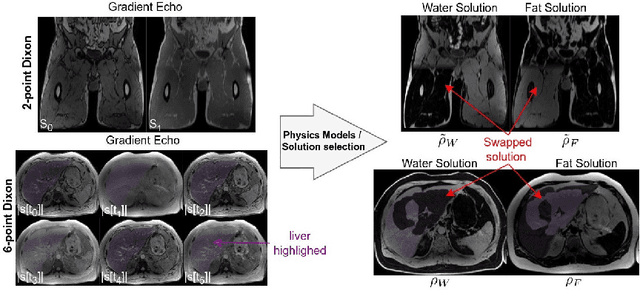
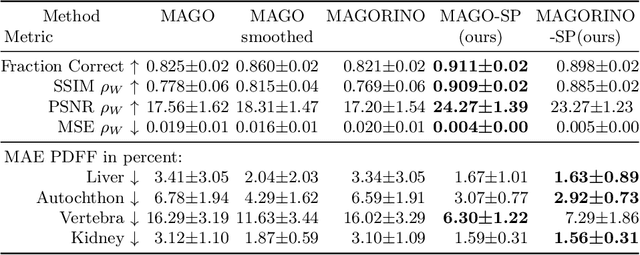
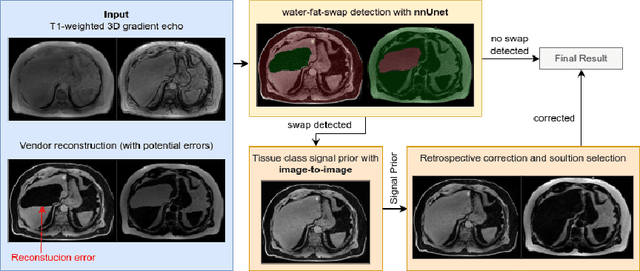
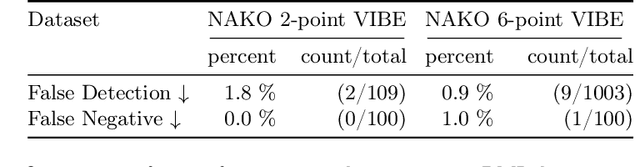
Abstract:Volume Interpolated Breath-Hold Examination (VIBE) MRI generates images suitable for water and fat signal composition estimation. While the two-point VIBE provides water-fat-separated images, the six-point VIBE allows estimation of the effective transversal relaxation rate R2* and the proton density fat fraction (PDFF), which are imaging markers for health and disease. Ambiguity during signal reconstruction can lead to water-fat swaps. This shortcoming challenges the application of VIBE-MRI for automated PDFF analyses of large-scale clinical data and of population studies. This study develops an automated pipeline to detect and correct water-fat swaps in non-contrast-enhanced VIBE images. Our three-step pipeline begins with training a segmentation network to classify volumes as "fat-like" or "water-like," using synthetic water-fat swaps generated by merging fat and water volumes with Perlin noise. Next, a denoising diffusion image-to-image network predicts water volumes as signal priors for correction. Finally, we integrate this prior into a physics-constrained model to recover accurate water and fat signals. Our approach achieves a < 1% error rate in water-fat swap detection for a 6-point VIBE. Notably, swaps disproportionately affect individuals in the Underweight and Class 3 Obesity BMI categories. Our correction algorithm ensures accurate solution selection in chemical phase MRIs, enabling reliable PDFF estimation. This forms a solid technical foundation for automated large-scale population imaging analysis.
Detecting Unforeseen Data Properties with Diffusion Autoencoder Embeddings using Spine MRI data
Oct 14, 2024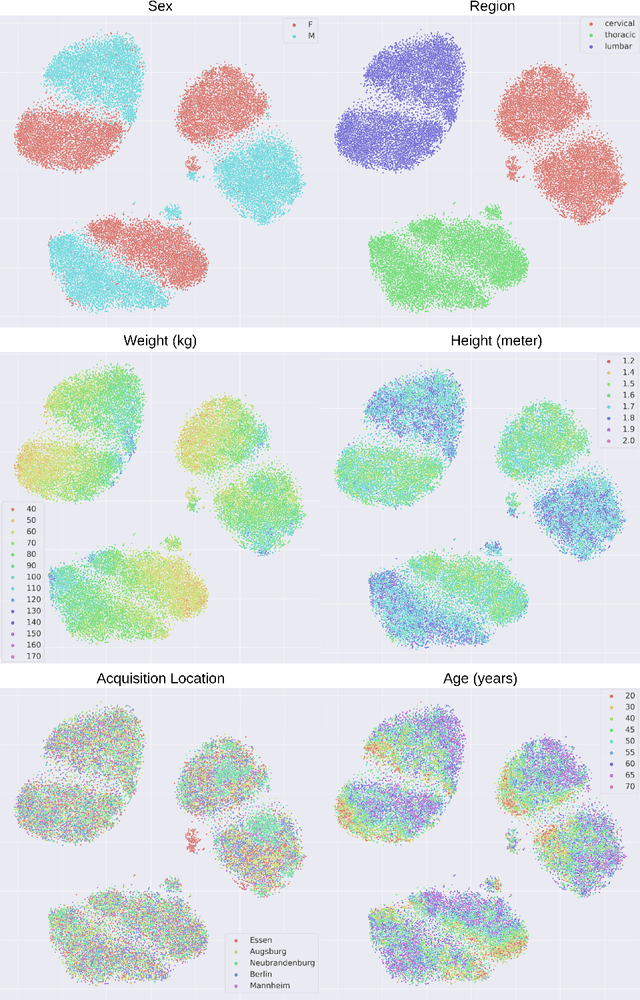

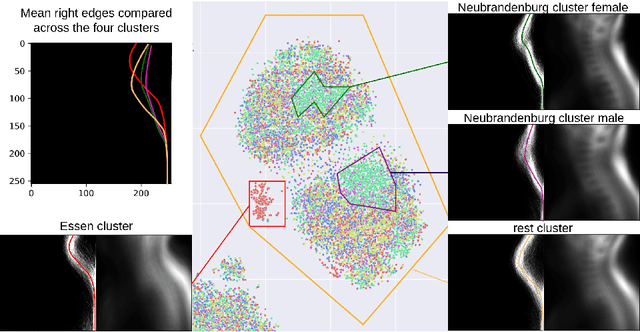
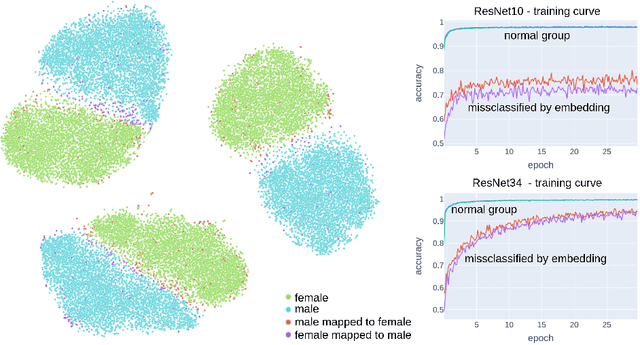
Abstract:Deep learning has made significant strides in medical imaging, leveraging the use of large datasets to improve diagnostics and prognostics. However, large datasets often come with inherent errors through subject selection and acquisition. In this paper, we investigate the use of Diffusion Autoencoder (DAE) embeddings for uncovering and understanding data characteristics and biases, including biases for protected variables like sex and data abnormalities indicative of unwanted protocol variations. We use sagittal T2-weighted magnetic resonance (MR) images of the neck, chest, and lumbar region from 11186 German National Cohort (NAKO) participants. We compare DAE embeddings with existing generative models like StyleGAN and Variational Autoencoder. Evaluations on a large-scale dataset consisting of sagittal T2-weighted MR images of three spine regions show that DAE embeddings effectively separate protected variables such as sex and age. Furthermore, we used t-SNE visualization to identify unwanted variations in imaging protocols, revealing differences in head positioning. Our embedding can identify samples where a sex predictor will have issues learning the correct sex. Our findings highlight the potential of using advanced embedding techniques like DAEs to detect data quality issues and biases in medical imaging datasets. Identifying such hidden relations can enhance the reliability and fairness of deep learning models in healthcare applications, ultimately improving patient care and outcomes.
Whole Heart 3D+T Representation Learning Through Sparse 2D Cardiac MR Images
Jun 01, 2024



Abstract:Cardiac Magnetic Resonance (CMR) imaging serves as the gold-standard for evaluating cardiac morphology and function. Typically, a multi-view CMR stack, covering short-axis (SA) and 2/3/4-chamber long-axis (LA) views, is acquired for a thorough cardiac assessment. However, efficiently streamlining the complex, high-dimensional 3D+T CMR data and distilling compact, coherent representation remains a challenge. In this work, we introduce a whole-heart self-supervised learning framework that utilizes masked imaging modeling to automatically uncover the correlations between spatial and temporal patches throughout the cardiac stacks. This process facilitates the generation of meaningful and well-clustered heart representations without relying on the traditionally required, and often costly, labeled data. The learned heart representation can be directly used for various downstream tasks. Furthermore, our method demonstrates remarkable robustness, ensuring consistent representations even when certain CMR planes are missing/flawed. We train our model on 14,000 unlabeled CMR data from UK BioBank and evaluate it on 1,000 annotated data. The proposed method demonstrates superior performance to baselines in tasks that demand comprehensive 3D+T cardiac information, e.g. cardiac phenotype (ejection fraction and ventricle volume) prediction and multi-plane/multi-frame CMR segmentation, highlighting its effectiveness in extracting comprehensive cardiac features that are both anatomically and pathologically relevant.
TotalVibeSegmentator: Full Torso Segmentation for the NAKO and UK Biobank in Volumetric Interpolated Breath-hold Examination Body Images
May 31, 2024



Abstract:Objectives: To present a publicly available torso segmentation network for large epidemiology datasets on volumetric interpolated breath-hold examination (VIBE) images. Materials & Methods: We extracted preliminary segmentations from TotalSegmentator, spine, and body composition networks for VIBE images, then improved them iteratively and retrained a nnUNet network. Using subsets of NAKO (85 subjects) and UK Biobank (16 subjects), we evaluated with Dice-score on a holdout set (12 subjects) and existing organ segmentation approach (1000 subjects), generating 71 semantic segmentation types for VIBE images. We provide an additional network for the vertebra segments 22 individual vertebra types. Results: We achieved an average Dice score of 0.89 +- 0.07 overall 71 segmentation labels. We scored > 0.90 Dice-score on the abdominal organs except for the pancreas with a Dice of 0.70. Conclusion: Our work offers a detailed and refined publicly available full torso segmentation on VIBE images.
Diff-Def: Diffusion-Generated Deformation Fields for Conditional Atlases
Mar 25, 2024



Abstract:Anatomical atlases are widely used for population analysis. Conditional atlases target a particular sub-population defined via certain conditions (e.g. demographics or pathologies) and allow for the investigation of fine-grained anatomical differences - such as morphological changes correlated with age. Existing approaches use either registration-based methods that are unable to handle large anatomical variations or generative models, which can suffer from training instabilities and hallucinations. To overcome these limitations, we use latent diffusion models to generate deformation fields, which transform a general population atlas into one representing a specific sub-population. By generating a deformation field and registering the conditional atlas to a neighbourhood of images, we ensure structural plausibility and avoid hallucinations, which can occur during direct image synthesis. We compare our method to several state-of-the-art atlas generation methods in experiments using 5000 brain as well as whole-body MR images from UK Biobank. Our method generates highly realistic atlases with smooth transformations and high anatomical fidelity, outperforming the baselines.
A Comparative Study of Population-Graph Construction Methods and Graph Neural Networks for Brain Age Regression
Sep 26, 2023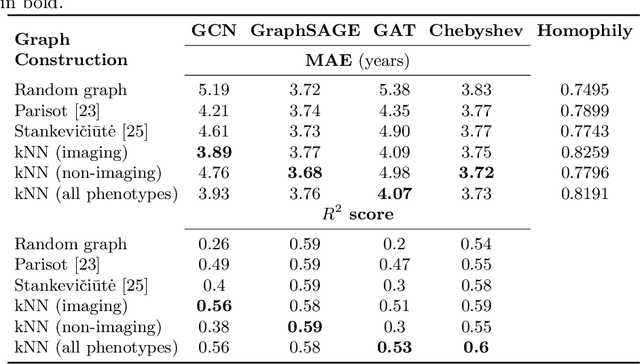
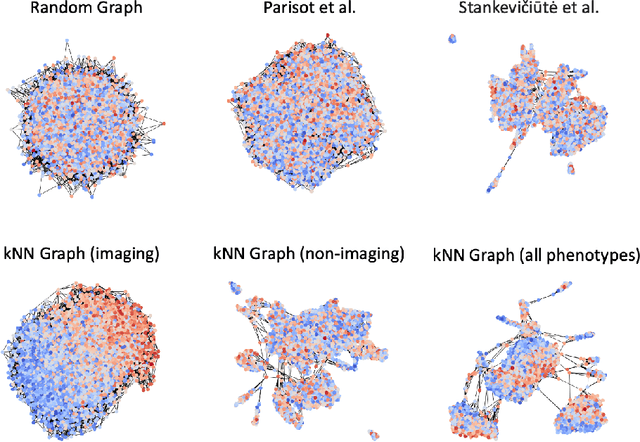
Abstract:The difference between the chronological and biological brain age of a subject can be an important biomarker for neurodegenerative diseases, thus brain age estimation can be crucial in clinical settings. One way to incorporate multimodal information into this estimation is through population graphs, which combine various types of imaging data and capture the associations among individuals within a population. In medical imaging, population graphs have demonstrated promising results, mostly for classification tasks. In most cases, the graph structure is pre-defined and remains static during training. However, extracting population graphs is a non-trivial task and can significantly impact the performance of Graph Neural Networks (GNNs), which are sensitive to the graph structure. In this work, we highlight the importance of a meaningful graph construction and experiment with different population-graph construction methods and their effect on GNN performance on brain age estimation. We use the homophily metric and graph visualizations to gain valuable quantitative and qualitative insights on the extracted graph structures. For the experimental evaluation, we leverage the UK Biobank dataset, which offers many imaging and non-imaging phenotypes. Our results indicate that architectures highly sensitive to the graph structure, such as Graph Convolutional Network (GCN) and Graph Attention Network (GAT), struggle with low homophily graphs, while other architectures, such as GraphSage and Chebyshev, are more robust across different homophily ratios. We conclude that static graph construction approaches are potentially insufficient for the task of brain age estimation and make recommendations for alternative research directions.
Constructing Population-Specific Atlases from Whole Body MRI: Application to the UKBB
Aug 28, 2023Abstract:Population atlases are commonly utilised in medical imaging to facilitate the investigation of variability across populations. Such atlases enable the mapping of medical images into a common coordinate system, promoting comparability and enabling the study of inter-subject differences. Constructing such atlases becomes particularly challenging when working with highly heterogeneous datasets, such as whole-body images, where subjects show significant anatomical variations. In this work, we propose a pipeline for generating a standardised whole-body atlas for a highly heterogeneous population by partitioning the population into meaningful subgroups. We create six whole-body atlases that represent a healthy population average using magnetic resonance (MR) images from the UK Biobank dataset. We furthermore unbias them, and this way obtain a realistic representation of the population. In addition to the anatomical atlases, we generate probabilistic atlases that capture the distributions of abdominal fat and five abdominal organs across the population. We demonstrate different applications of these atlases, using the differences between subjects with medical conditions such as diabetes and cardiovascular diseases and healthy subjects from the atlas space. With this work, we make the constructed anatomical and label atlases publically available and anticipate them to support medical research conducted on whole-body MR images.
Unlocking the Diagnostic Potential of ECG through Knowledge Transfer from Cardiac MRI
Aug 09, 2023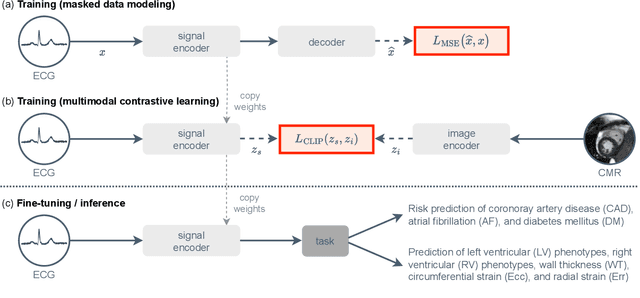
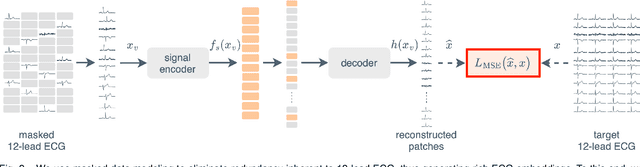
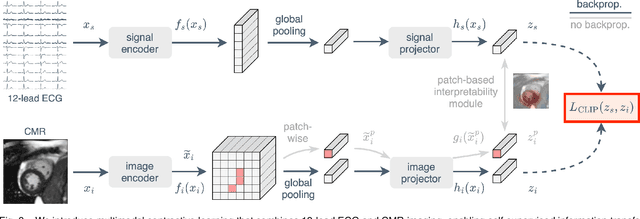
Abstract:The electrocardiogram (ECG) is a widely available diagnostic tool that allows for a cost-effective and fast assessment of the cardiovascular health. However, more detailed examination with expensive cardiac magnetic resonance (CMR) imaging is often preferred for the diagnosis of cardiovascular diseases. While providing detailed visualization of the cardiac anatomy, CMR imaging is not widely available due to long scan times and high costs. To address this issue, we propose the first self-supervised contrastive approach that transfers domain-specific information from CMR images to ECG embeddings. Our approach combines multimodal contrastive learning with masked data modeling to enable holistic cardiac screening solely from ECG data. In extensive experiments using data from 40,044 UK Biobank subjects, we demonstrate the utility and generalizability of our method. We predict the subject-specific risk of various cardiovascular diseases and determine distinct cardiac phenotypes solely from ECG data. In a qualitative analysis, we demonstrate that our learned ECG embeddings incorporate information from CMR image regions of interest. We make our entire pipeline publicly available, including the source code and pre-trained model weights.
Atlas-Based Interpretable Age Prediction
Jul 14, 2023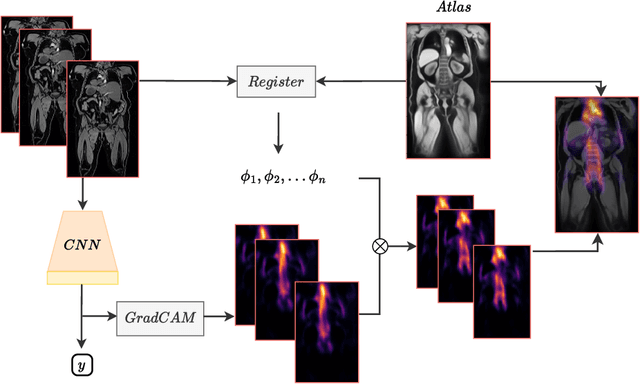
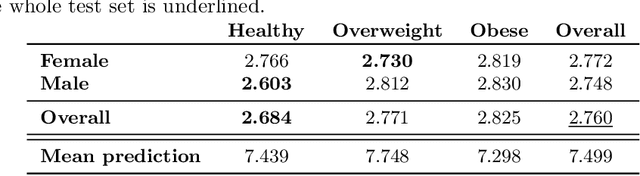
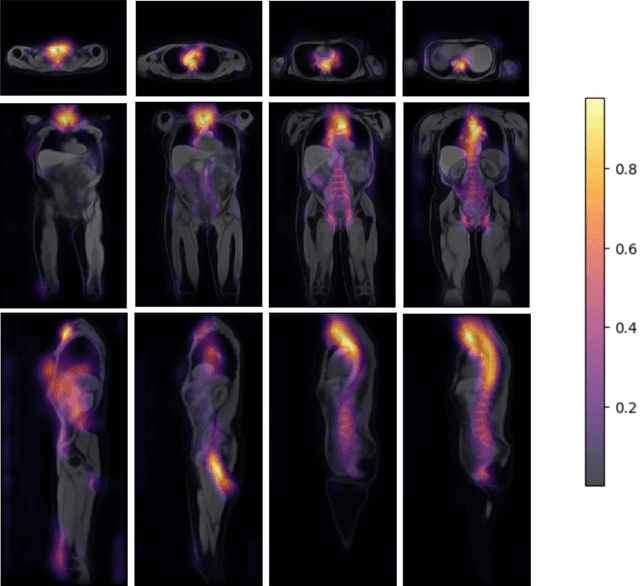
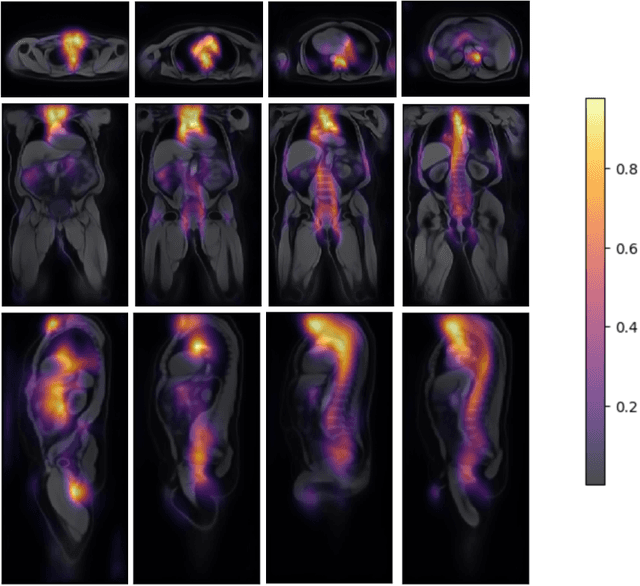
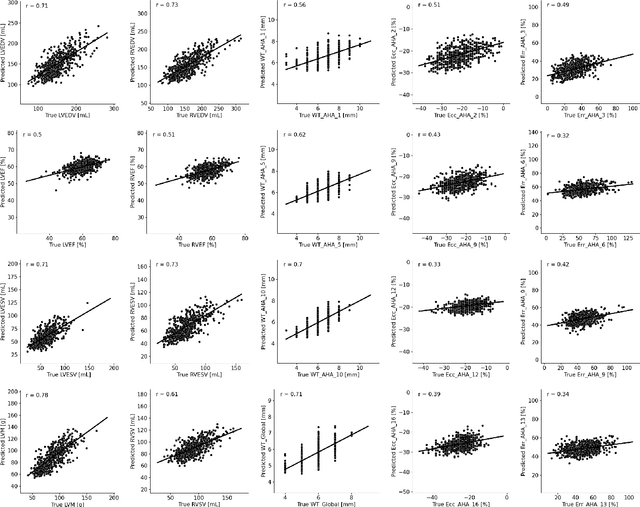
Abstract:Age prediction is an important part of medical assessments and research. It can aid in detecting diseases as well as abnormal ageing by highlighting the discrepancy between chronological and biological age. To gain a comprehensive understanding of age-related changes observed in various body parts, we investigate them on a larger scale by using whole-body images. We utilise the Grad-CAM interpretability method to determine the body areas most predictive of a person's age. We expand our analysis beyond individual subjects by employing registration techniques to generate population-wide interpretability maps. Furthermore, we set state-of-the-art whole-body age prediction with a model that achieves a mean absolute error of 2.76 years. Our findings reveal three primary areas of interest: the spine, the autochthonous back muscles, and the cardiac region, which exhibits the highest importance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge