Vasileios Baltatzis
Neural Sign Actors: A diffusion model for 3D sign language production from text
Dec 05, 2023Abstract:Sign Languages (SL) serve as the predominant mode of communication for the Deaf and Hard of Hearing communities. The advent of deep learning has aided numerous methods in SL recognition and translation, achieving remarkable results. However, Sign Language Production (SLP) poses a challenge for the computer vision community as the motions generated must be realistic and have precise semantic meanings. Most SLP methods rely on 2D data, thus impeding their ability to attain a necessary level of realism. In this work, we propose a diffusion-based SLP model trained on a curated large-scale dataset of 4D signing avatars and their corresponding text transcripts. The proposed method can generate dynamic sequences of 3D avatars from an unconstrained domain of discourse using a diffusion process formed on a novel and anatomically informed graph neural network defined on the SMPL-X body skeleton. Through a series of quantitative and qualitative experiments, we show that the proposed method considerably outperforms previous methods of SLP. We believe that this work presents an important and necessary step towards realistic neural sign avatars, bridging the communication gap between Deaf and hearing communities. The code, method and generated data will be made publicly available.
KGEx: Explaining Knowledge Graph Embeddings via Subgraph Sampling and Knowledge Distillation
Oct 02, 2023Abstract:Despite being the go-to choice for link prediction on knowledge graphs, research on interpretability of knowledge graph embeddings (KGE) has been relatively unexplored. We present KGEx, a novel post-hoc method that explains individual link predictions by drawing inspiration from surrogate models research. Given a target triple to predict, KGEx trains surrogate KGE models that we use to identify important training triples. To gauge the impact of a training triple, we sample random portions of the target triple neighborhood and we train multiple surrogate KGE models on each of them. To ensure faithfulness, each surrogate is trained by distilling knowledge from the original KGE model. We then assess how well surrogates predict the target triple being explained, the intuition being that those leading to faithful predictions have been trained on impactful neighborhood samples. Under this assumption, we then harvest triples that appear frequently across impactful neighborhoods. We conduct extensive experiments on two publicly available datasets, to demonstrate that KGEx is capable of providing explanations faithful to the black-box model.
A Comparative Study of Population-Graph Construction Methods and Graph Neural Networks for Brain Age Regression
Sep 26, 2023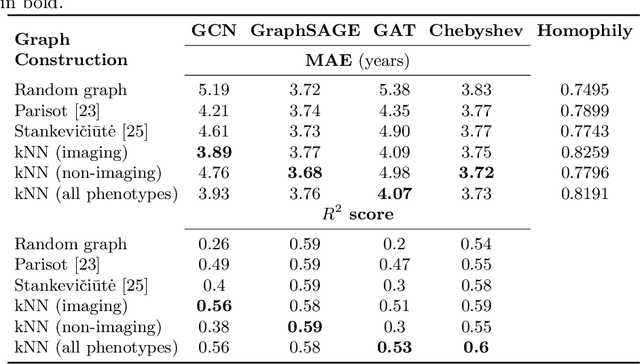
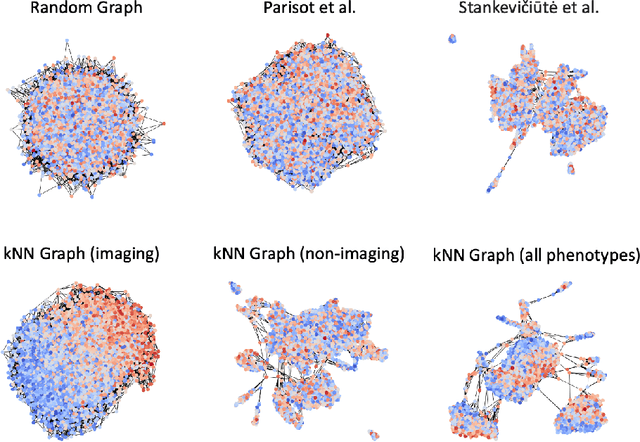
Abstract:The difference between the chronological and biological brain age of a subject can be an important biomarker for neurodegenerative diseases, thus brain age estimation can be crucial in clinical settings. One way to incorporate multimodal information into this estimation is through population graphs, which combine various types of imaging data and capture the associations among individuals within a population. In medical imaging, population graphs have demonstrated promising results, mostly for classification tasks. In most cases, the graph structure is pre-defined and remains static during training. However, extracting population graphs is a non-trivial task and can significantly impact the performance of Graph Neural Networks (GNNs), which are sensitive to the graph structure. In this work, we highlight the importance of a meaningful graph construction and experiment with different population-graph construction methods and their effect on GNN performance on brain age estimation. We use the homophily metric and graph visualizations to gain valuable quantitative and qualitative insights on the extracted graph structures. For the experimental evaluation, we leverage the UK Biobank dataset, which offers many imaging and non-imaging phenotypes. Our results indicate that architectures highly sensitive to the graph structure, such as Graph Convolutional Network (GCN) and Graph Attention Network (GAT), struggle with low homophily graphs, while other architectures, such as GraphSage and Chebyshev, are more robust across different homophily ratios. We conclude that static graph construction approaches are potentially insufficient for the task of brain age estimation and make recommendations for alternative research directions.
Multimodal brain age estimation using interpretable adaptive population-graph learning
Jul 19, 2023

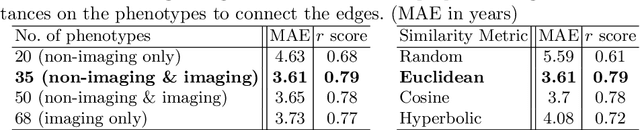

Abstract:Brain age estimation is clinically important as it can provide valuable information in the context of neurodegenerative diseases such as Alzheimer's. Population graphs, which include multimodal imaging information of the subjects along with the relationships among the population, have been used in literature along with Graph Convolutional Networks (GCNs) and have proved beneficial for a variety of medical imaging tasks. A population graph is usually static and constructed manually using non-imaging information. However, graph construction is not a trivial task and might significantly affect the performance of the GCN, which is inherently very sensitive to the graph structure. In this work, we propose a framework that learns a population graph structure optimized for the downstream task. An attention mechanism assigns weights to a set of imaging and non-imaging features (phenotypes), which are then used for edge extraction. The resulting graph is used to train the GCN. The entire pipeline can be trained end-to-end. Additionally, by visualizing the attention weights that were the most important for the graph construction, we increase the interpretability of the graph. We use the UK Biobank, which provides a large variety of neuroimaging and non-imaging phenotypes, to evaluate our method on brain age regression and classification. The proposed method outperforms competing static graph approaches and other state-of-the-art adaptive methods. We further show that the assigned attention scores indicate that there are both imaging and non-imaging phenotypes that are informative for brain age estimation and are in agreement with the relevant literature.
Evaluation of 3D GANs for Lung Tissue Modelling in Pulmonary CT
Aug 17, 2022
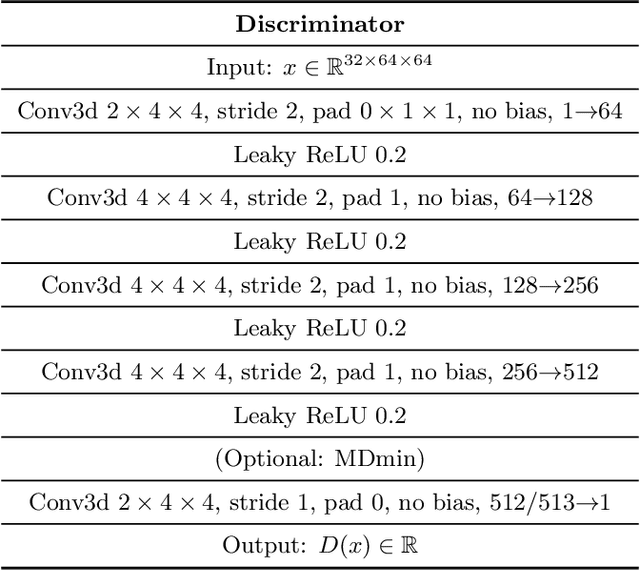
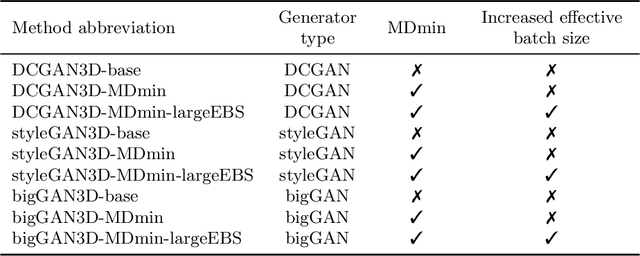

Abstract:GANs are able to model accurately the distribution of complex, high-dimensional datasets, e.g. images. This makes high-quality GANs useful for unsupervised anomaly detection in medical imaging. However, differences in training datasets such as output image dimensionality and appearance of semantically meaningful features mean that GAN models from the natural image domain may not work `out-of-the-box' for medical imaging, necessitating re-implementation and re-evaluation. In this work we adapt and evaluate three GAN models to the task of modelling 3D healthy image patches for pulmonary CT. To the best of our knowledge, this is the first time that such an evaluation has been performed. The DCGAN, styleGAN and the bigGAN architectures were investigated due to their ubiquity and high performance in natural image processing. We train different variants of these methods and assess their performance using the FID score. In addition, the quality of the generated images was evaluated by a human observer study, the ability of the networks to model 3D domain-specific features was investigated, and the structure of the GAN latent spaces was analysed. Results show that the 3D styleGAN produces realistic-looking images with meaningful 3D structure, but suffer from mode collapse which must be addressed during training to obtain samples diversity. Conversely, the 3D DCGAN models show a greater capacity for image variability, but at the cost of poor-quality images. The 3D bigGAN models provide an intermediate level of image quality, but most accurately model the distribution of selected semantically meaningful features. The results suggest that future development is required to realise a 3D GAN with sufficient capacity for patch-based lung CT anomaly detection and we offer recommendations for future areas of research, such as experimenting with other architectures and incorporation of position-encoding.
Is MC Dropout Bayesian?
Oct 08, 2021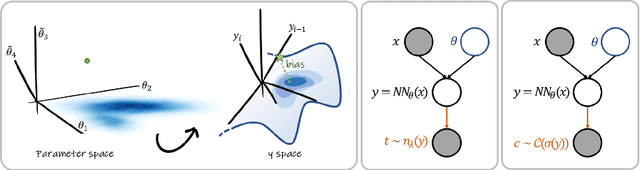

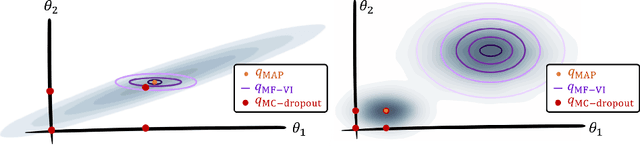

Abstract:MC Dropout is a mainstream "free lunch" method in medical imaging for approximate Bayesian computations (ABC). Its appeal is to solve out-of-the-box the daunting task of ABC and uncertainty quantification in Neural Networks (NNs); to fall within the variational inference (VI) framework; and to propose a highly multimodal, faithful predictive posterior. We question the properties of MC Dropout for approximate inference, as in fact MC Dropout changes the Bayesian model; its predictive posterior assigns $0$ probability to the true model on closed-form benchmarks; the multimodality of its predictive posterior is not a property of the true predictive posterior but a design artefact. To address the need for VI on arbitrary models, we share a generic VI engine within the pytorch framework. The code includes a carefully designed implementation of structured (diagonal plus low-rank) multivariate normal variational families, and mixtures thereof. It is intended as a go-to no-free-lunch approach, addressing shortcomings of mean-field VI with an adjustable trade-off between expressivity and computational complexity.
Voxel-level Importance Maps for Interpretable Brain Age Estimation
Aug 11, 2021

Abstract:Brain aging, and more specifically the difference between the chronological and the biological age of a person, may be a promising biomarker for identifying neurodegenerative diseases. For this purpose accurate prediction is important but the localisation of the areas that play a significant role in the prediction is also crucial, in order to gain clinicians' trust and reassurance about the performance of a prediction model. Most interpretability methods are focused on classification tasks and cannot be directly transferred to regression tasks. In this study, we focus on the task of brain age regression from 3D brain Magnetic Resonance (MR) images using a Convolutional Neural Network, termed prediction model. We interpret its predictions by extracting importance maps, which discover the parts of the brain that are the most important for brain age. In order to do so, we assume that voxels that are not useful for the regression are resilient to noise addition. We implement a noise model which aims to add as much noise as possible to the input without harming the performance of the prediction model. We average the importance maps of the subjects and end up with a population-based importance map, which displays the regions of the brain that are influential for the task. We test our method on 13,750 3D brain MR images from the UK Biobank, and our findings are consistent with the existing neuropathology literature, highlighting that the hippocampus and the ventricles are the most relevant regions for brain aging.
The Pitfalls of Sample Selection: A Case Study on Lung Nodule Classification
Aug 11, 2021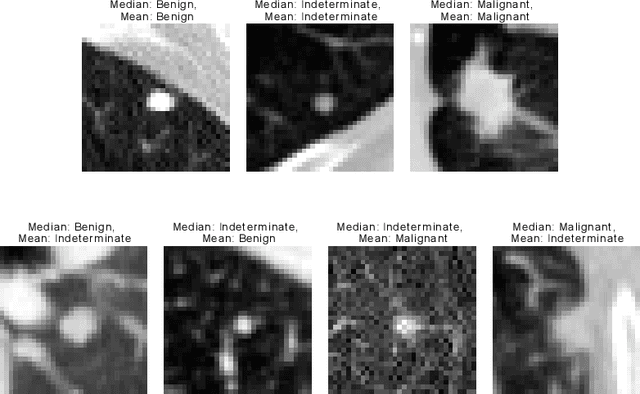
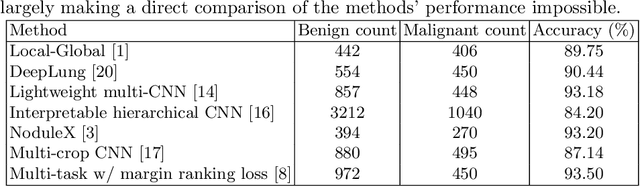
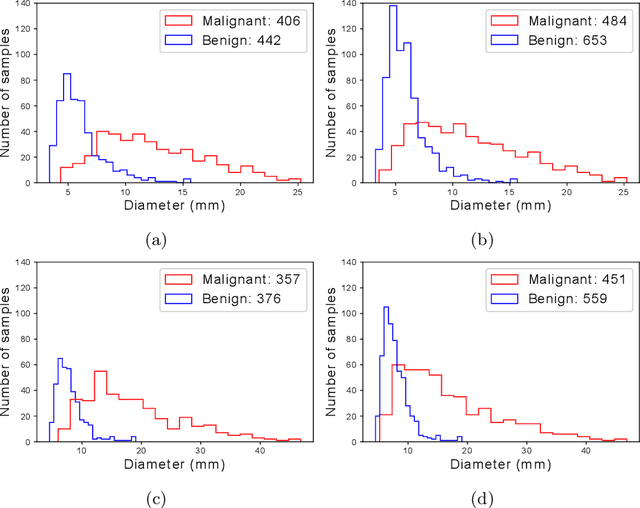
Abstract:Using publicly available data to determine the performance of methodological contributions is important as it facilitates reproducibility and allows scrutiny of the published results. In lung nodule classification, for example, many works report results on the publicly available LIDC dataset. In theory, this should allow a direct comparison of the performance of proposed methods and assess the impact of individual contributions. When analyzing seven recent works, however, we find that each employs a different data selection process, leading to largely varying total number of samples and ratios between benign and malignant cases. As each subset will have different characteristics with varying difficulty for classification, a direct comparison between the proposed methods is thus not always possible, nor fair. We study the particular effect of truthing when aggregating labels from multiple experts. We show that specific choices can have severe impact on the data distribution where it may be possible to achieve superior performance on one sample distribution but not on another. While we show that we can further improve on the state-of-the-art on one sample selection, we also find that on a more challenging sample selection, on the same database, the more advanced models underperform with respect to very simple baseline methods, highlighting that the selected data distribution may play an even more important role than the model architecture. This raises concerns about the validity of claimed methodological contributions. We believe the community should be aware of these pitfalls and make recommendations on how these can be avoided in future work.
The Effect of the Loss on Generalization: Empirical Study on Synthetic Lung Nodule Data
Aug 10, 2021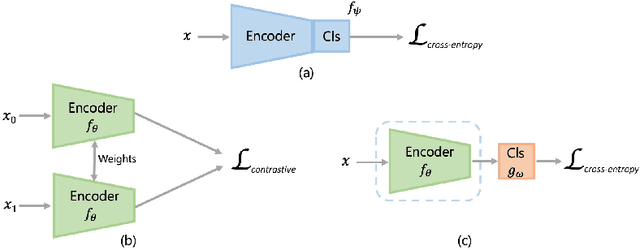
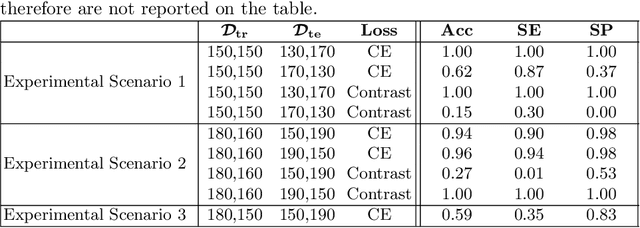
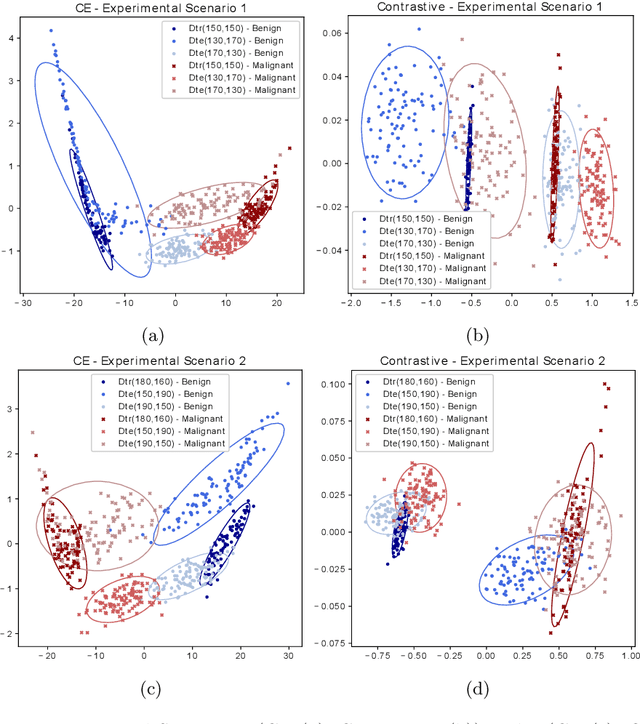
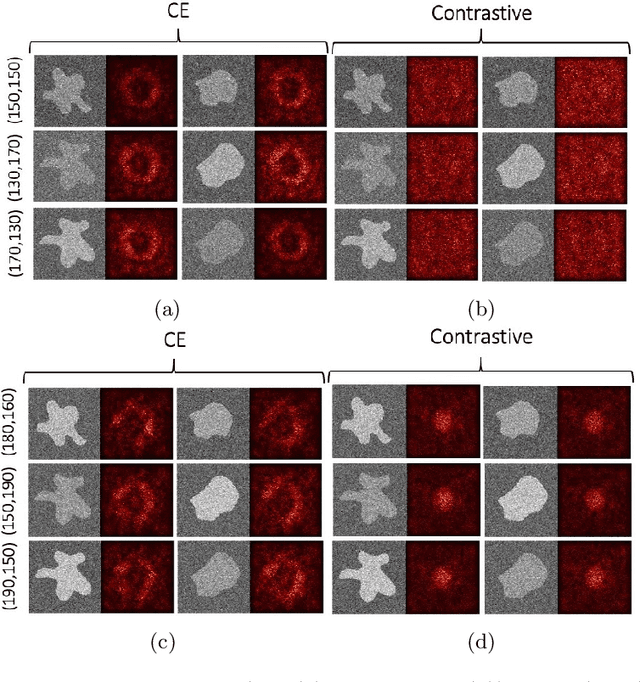
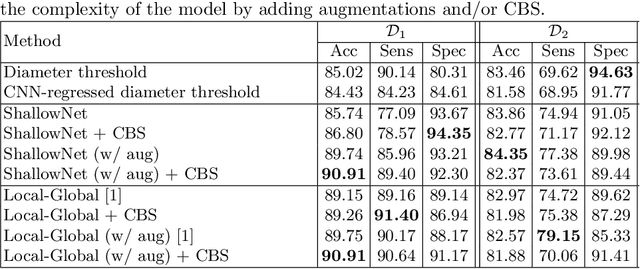
Abstract:Convolutional Neural Networks (CNNs) are widely used for image classification in a variety of fields, including medical imaging. While most studies deploy cross-entropy as the loss function in such tasks, a growing number of approaches have turned to a family of contrastive learning-based losses. Even though performance metrics such as accuracy, sensitivity and specificity are regularly used for the evaluation of CNN classifiers, the features that these classifiers actually learn are rarely identified and their effect on the classification performance on out-of-distribution test samples is insufficiently explored. In this paper, motivated by the real-world task of lung nodule classification, we investigate the features that a CNN learns when trained and tested on different distributions of a synthetic dataset with controlled modes of variation. We show that different loss functions lead to different features being learned and consequently affect the generalization ability of the classifier on unseen data. This study provides some important insights into the design of deep learning solutions for medical imaging tasks.
Bayesian analysis of the prevalence bias: learning and predicting from imbalanced data
Jul 31, 2021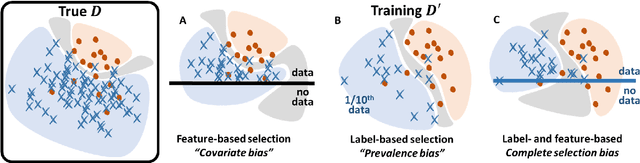
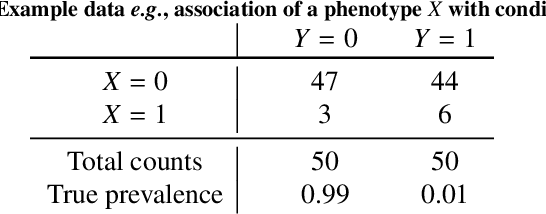

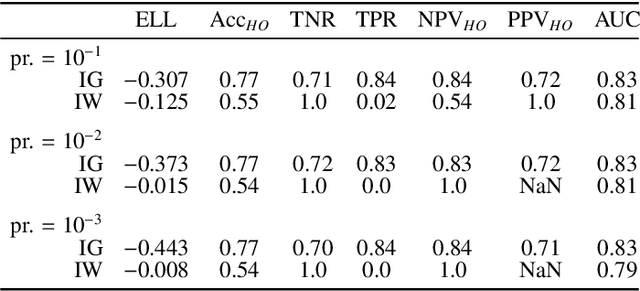
Abstract:Datasets are rarely a realistic approximation of the target population. Say, prevalence is misrepresented, image quality is above clinical standards, etc. This mismatch is known as sampling bias. Sampling biases are a major hindrance for machine learning models. They cause significant gaps between model performance in the lab and in the real world. Our work is a solution to prevalence bias. Prevalence bias is the discrepancy between the prevalence of a pathology and its sampling rate in the training dataset, introduced upon collecting data or due to the practioner rebalancing the training batches. This paper lays the theoretical and computational framework for training models, and for prediction, in the presence of prevalence bias. Concretely a bias-corrected loss function, as well as bias-corrected predictive rules, are derived under the principles of Bayesian risk minimization. The loss exhibits a direct connection to the information gain. It offers a principled alternative to heuristic training losses and complements test-time procedures based on selecting an operating point from summary curves. It integrates seamlessly in the current paradigm of (deep) learning using stochastic backpropagation and naturally with Bayesian models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge