Tamara T. Mueller
Parametric shape models for vessels learned from segmentations via differentiable voxelization
Jul 03, 2025Abstract:Vessels are complex structures in the body that have been studied extensively in multiple representations. While voxelization is the most common of them, meshes and parametric models are critical in various applications due to their desirable properties. However, these representations are typically extracted through segmentations and used disjointly from each other. We propose a framework that joins the three representations under differentiable transformations. By leveraging differentiable voxelization, we automatically extract a parametric shape model of the vessels through shape-to-segmentation fitting, where we learn shape parameters from segmentations without the explicit need for ground-truth shape parameters. The vessel is parametrized as centerlines and radii using cubic B-splines, ensuring smoothness and continuity by construction. Meshes are differentiably extracted from the learned shape parameters, resulting in high-fidelity meshes that can be manipulated post-fit. Our method can accurately capture the geometry of complex vessels, as demonstrated by the volumetric fits in experiments on aortas, aneurysms, and brain vessels.
Bounding Reconstruction Attack Success of Adversaries Without Data Priors
Feb 20, 2024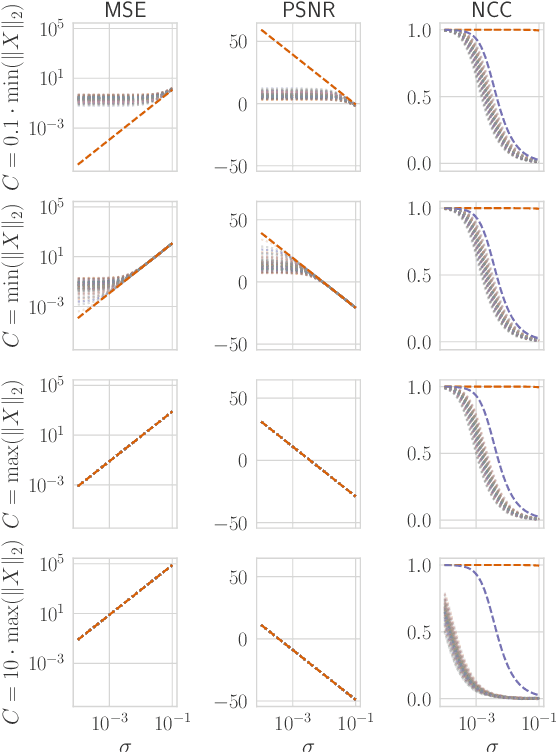
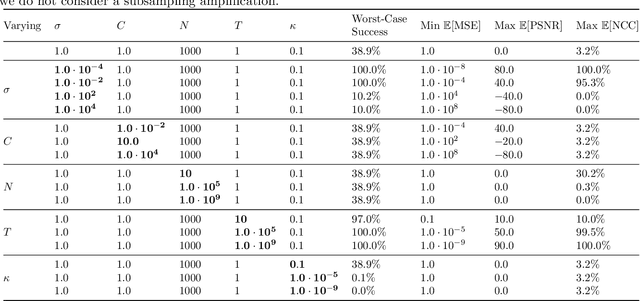
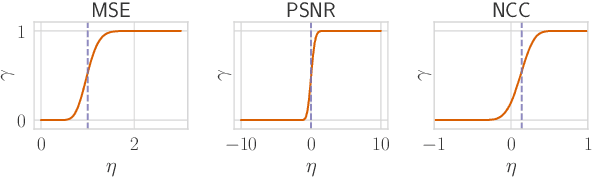
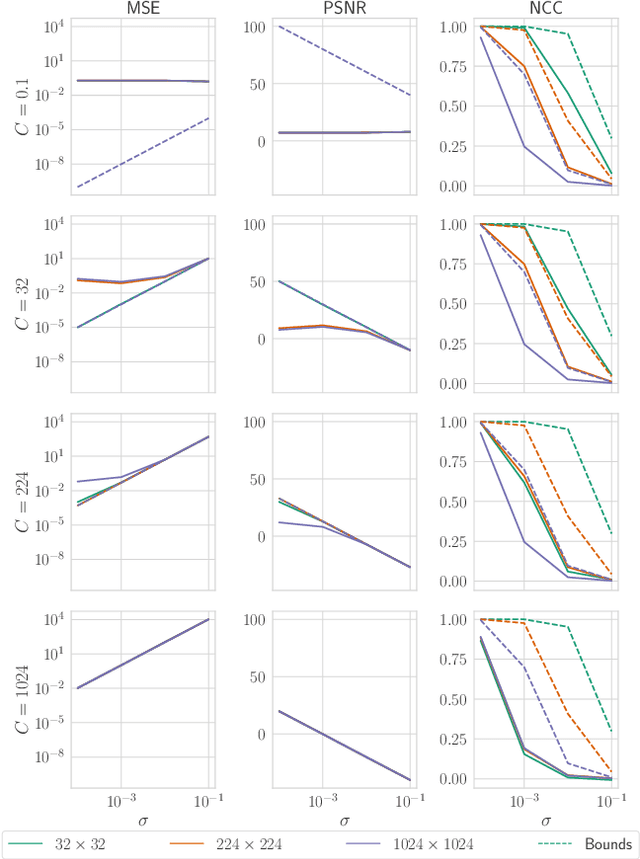
Abstract:Reconstruction attacks on machine learning (ML) models pose a strong risk of leakage of sensitive data. In specific contexts, an adversary can (almost) perfectly reconstruct training data samples from a trained model using the model's gradients. When training ML models with differential privacy (DP), formal upper bounds on the success of such reconstruction attacks can be provided. So far, these bounds have been formulated under worst-case assumptions that might not hold high realistic practicality. In this work, we provide formal upper bounds on reconstruction success under realistic adversarial settings against ML models trained with DP and support these bounds with empirical results. With this, we show that in realistic scenarios, (a) the expected reconstruction success can be bounded appropriately in different contexts and by different metrics, which (b) allows for a more educated choice of a privacy parameter.
Reconciling AI Performance and Data Reconstruction Resilience for Medical Imaging
Dec 05, 2023

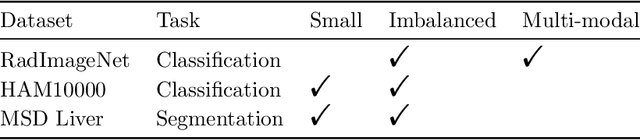

Abstract:Artificial Intelligence (AI) models are vulnerable to information leakage of their training data, which can be highly sensitive, for example in medical imaging. Privacy Enhancing Technologies (PETs), such as Differential Privacy (DP), aim to circumvent these susceptibilities. DP is the strongest possible protection for training models while bounding the risks of inferring the inclusion of training samples or reconstructing the original data. DP achieves this by setting a quantifiable privacy budget. Although a lower budget decreases the risk of information leakage, it typically also reduces the performance of such models. This imposes a trade-off between robust performance and stringent privacy. Additionally, the interpretation of a privacy budget remains abstract and challenging to contextualize. In this study, we contrast the performance of AI models at various privacy budgets against both, theoretical risk bounds and empirical success of reconstruction attacks. We show that using very large privacy budgets can render reconstruction attacks impossible, while drops in performance are negligible. We thus conclude that not using DP -- at all -- is negligent when applying AI models to sensitive data. We deem those results to lie a foundation for further debates on striking a balance between privacy risks and model performance.
A Comparative Study of Population-Graph Construction Methods and Graph Neural Networks for Brain Age Regression
Sep 26, 2023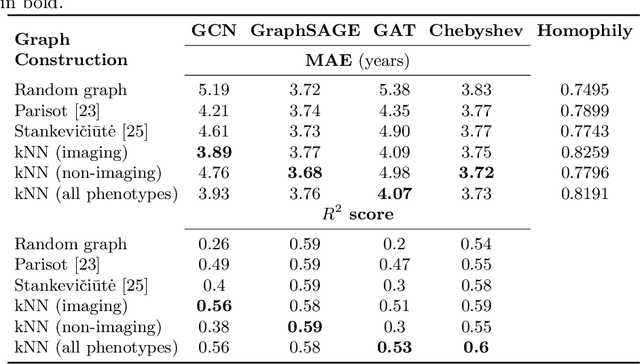
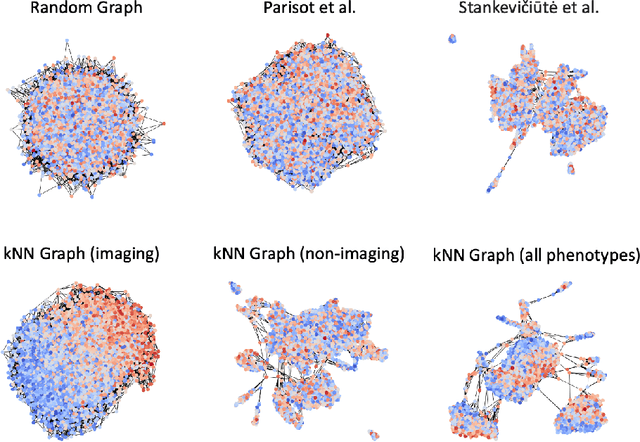
Abstract:The difference between the chronological and biological brain age of a subject can be an important biomarker for neurodegenerative diseases, thus brain age estimation can be crucial in clinical settings. One way to incorporate multimodal information into this estimation is through population graphs, which combine various types of imaging data and capture the associations among individuals within a population. In medical imaging, population graphs have demonstrated promising results, mostly for classification tasks. In most cases, the graph structure is pre-defined and remains static during training. However, extracting population graphs is a non-trivial task and can significantly impact the performance of Graph Neural Networks (GNNs), which are sensitive to the graph structure. In this work, we highlight the importance of a meaningful graph construction and experiment with different population-graph construction methods and their effect on GNN performance on brain age estimation. We use the homophily metric and graph visualizations to gain valuable quantitative and qualitative insights on the extracted graph structures. For the experimental evaluation, we leverage the UK Biobank dataset, which offers many imaging and non-imaging phenotypes. Our results indicate that architectures highly sensitive to the graph structure, such as Graph Convolutional Network (GCN) and Graph Attention Network (GAT), struggle with low homophily graphs, while other architectures, such as GraphSage and Chebyshev, are more robust across different homophily ratios. We conclude that static graph construction approaches are potentially insufficient for the task of brain age estimation and make recommendations for alternative research directions.
Constructing Population-Specific Atlases from Whole Body MRI: Application to the UKBB
Aug 28, 2023Abstract:Population atlases are commonly utilised in medical imaging to facilitate the investigation of variability across populations. Such atlases enable the mapping of medical images into a common coordinate system, promoting comparability and enabling the study of inter-subject differences. Constructing such atlases becomes particularly challenging when working with highly heterogeneous datasets, such as whole-body images, where subjects show significant anatomical variations. In this work, we propose a pipeline for generating a standardised whole-body atlas for a highly heterogeneous population by partitioning the population into meaningful subgroups. We create six whole-body atlases that represent a healthy population average using magnetic resonance (MR) images from the UK Biobank dataset. We furthermore unbias them, and this way obtain a realistic representation of the population. In addition to the anatomical atlases, we generate probabilistic atlases that capture the distributions of abdominal fat and five abdominal organs across the population. We demonstrate different applications of these atlases, using the differences between subjects with medical conditions such as diabetes and cardiovascular diseases and healthy subjects from the atlas space. With this work, we make the constructed anatomical and label atlases publically available and anticipate them to support medical research conducted on whole-body MR images.
Extended Graph Assessment Metrics for Graph Neural Networks
Jul 13, 2023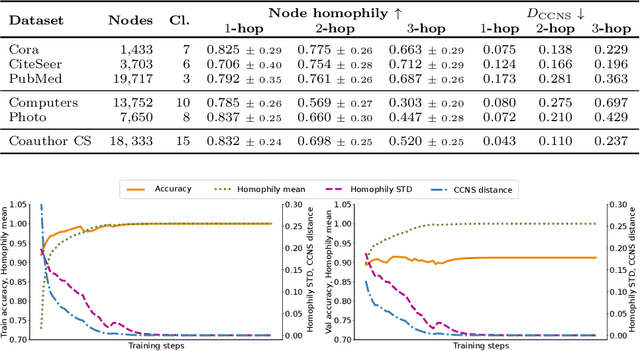
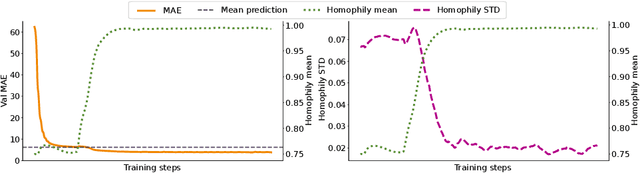
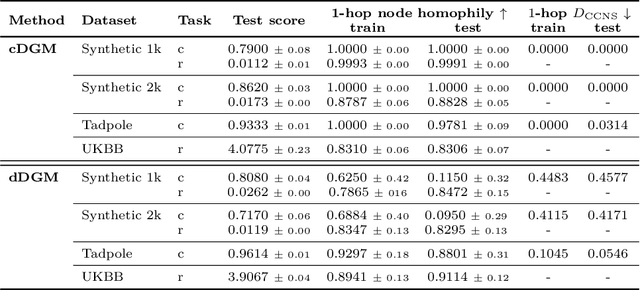
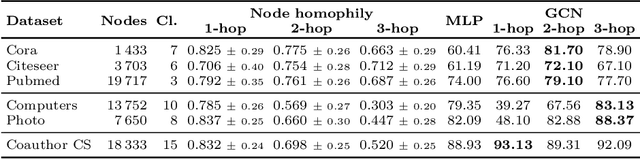
Abstract:When re-structuring patient cohorts into so-called population graphs, initially independent data points can be incorporated into one interconnected graph structure. This population graph can then be used for medical downstream tasks using graph neural networks (GNNs). The construction of a suitable graph structure is a challenging step in the learning pipeline that can have severe impact on model performance. To this end, different graph assessment metrics have been introduced to evaluate graph structures. However, these metrics are limited to classification tasks and discrete adjacency matrices, only covering a small subset of real-world applications. In this work, we introduce extended graph assessment metrics (GAMs) for regression tasks and continuous adjacency matrices. We focus on two GAMs in specific: \textit{homophily} and \textit{cross-class neighbourhood similarity} (CCNS). We extend the notion of GAMs to more than one hop, define homophily for regression tasks, as well as continuous adjacency matrices, and propose a light-weight CCNS distance for discrete and continuous adjacency matrices. We show the correlation of these metrics with model performance on different medical population graphs and under different learning settings.
Body Fat Estimation from Surface Meshes using Graph Neural Networks
Jul 13, 2023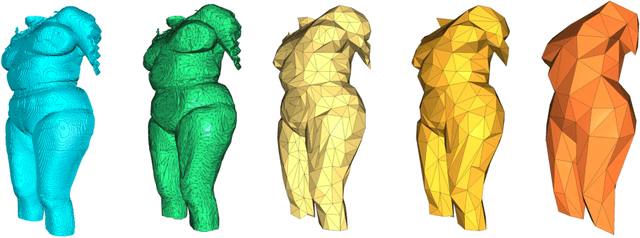

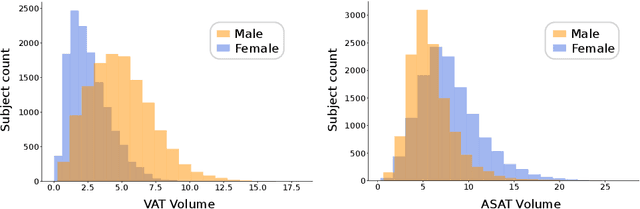

Abstract:Body fat volume and distribution can be a strong indication for a person's overall health and the risk for developing diseases like type 2 diabetes and cardiovascular diseases. Frequently used measures for fat estimation are the body mass index (BMI), waist circumference, or the waist-hip-ratio. However, those are rather imprecise measures that do not allow for a discrimination between different types of fat or between fat and muscle tissue. The estimation of visceral (VAT) and abdominal subcutaneous (ASAT) adipose tissue volume has shown to be a more accurate measure for named risk factors. In this work, we show that triangulated body surface meshes can be used to accurately predict VAT and ASAT volumes using graph neural networks. Our methods achieve high performance while reducing training time and required resources compared to state-of-the-art convolutional neural networks in this area. We furthermore envision this method to be applicable to cheaper and easily accessible medical surface scans instead of expensive medical images.
Privacy-Utility Trade-offs in Neural Networks for Medical Population Graphs: Insights from Differential Privacy and Graph Structure
Jul 13, 2023Abstract:We initiate an empirical investigation into differentially private graph neural networks on population graphs from the medical domain by examining privacy-utility trade-offs at different privacy levels on both real-world and synthetic datasets and performing auditing through membership inference attacks. Our findings highlight the potential and the challenges of this specific DP application area. Moreover, we find evidence that the underlying graph structure constitutes a potential factor for larger performance gaps by showing a correlation between the degree of graph homophily and the accuracy of the trained model.
Explainable 2D Vision Models for 3D Medical Data
Jul 13, 2023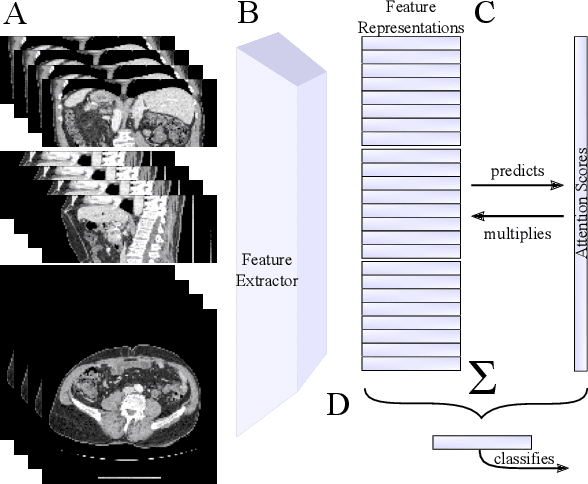
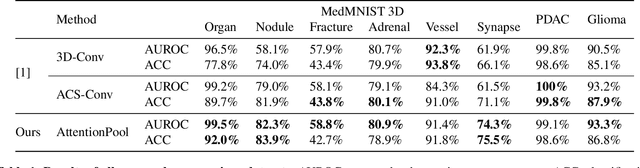
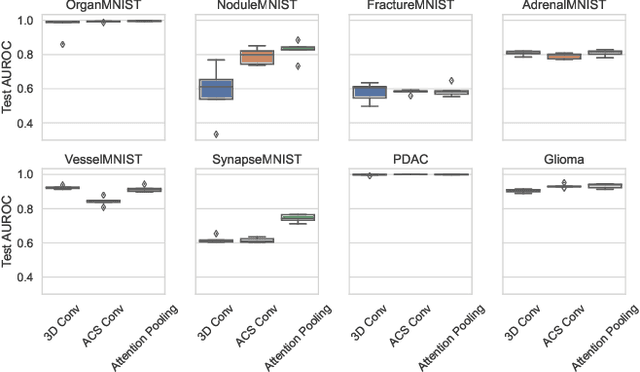
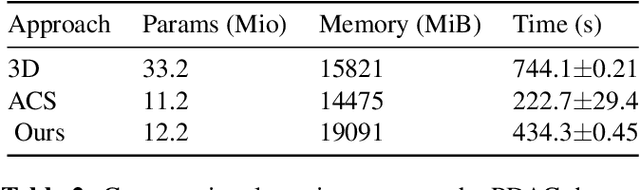
Abstract:Training Artificial Intelligence (AI) models on three-dimensional image data presents unique challenges compared to the two-dimensional case: Firstly, the computational resources are significantly higher, and secondly, the availability of large pretraining datasets is often limited, impeding training success. In this study, we propose a simple approach of adapting 2D networks with an intermediate feature representation for processing 3D volumes. Our method involves sequentially applying these networks to slices of a 3D volume from all orientations. Subsequently, a feature reduction module combines the extracted slice features into a single representation, which is then used for classification. We evaluate our approach on medical classification benchmarks and a real-world clinical dataset, demonstrating comparable results to existing methods. Furthermore, by employing attention pooling as a feature reduction module we obtain weighted importance values for each slice during the forward pass. We show that slices deemed important by our approach allow the inspection of the basis of a model's prediction.
How Do Input Attributes Impact the Privacy Loss in Differential Privacy?
Nov 18, 2022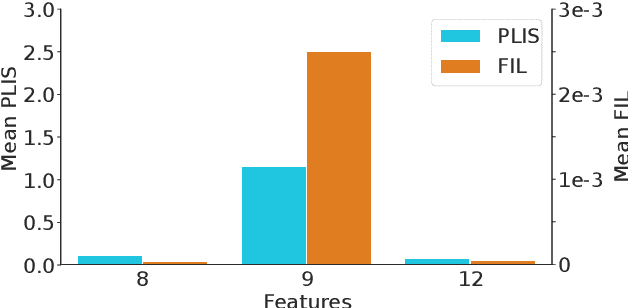

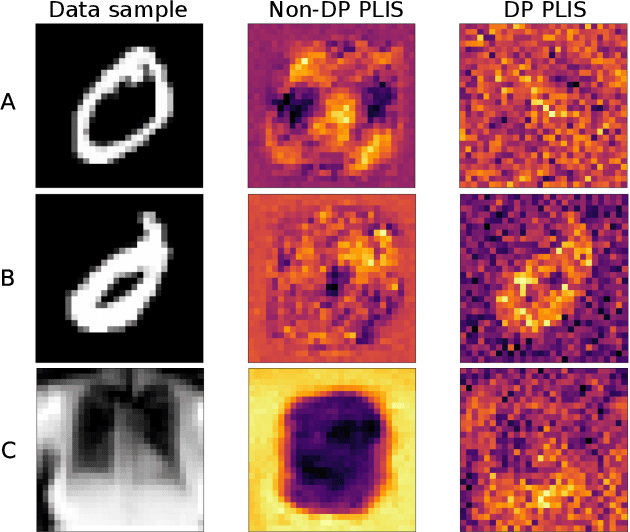
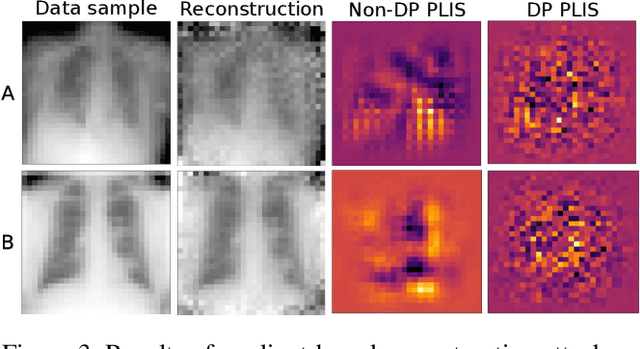
Abstract:Differential privacy (DP) is typically formulated as a worst-case privacy guarantee over all individuals in a database. More recently, extensions to individual subjects or their attributes, have been introduced. Under the individual/per-instance DP interpretation, we study the connection between the per-subject gradient norm in DP neural networks and individual privacy loss and introduce a novel metric termed the Privacy Loss-Input Susceptibility (PLIS), which allows one to apportion the subject's privacy loss to their input attributes. We experimentally show how this enables the identification of sensitive attributes and of subjects at high risk of data reconstruction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge