Kyriaki-Margarita Bintsi
Global 3D Reconstruction of Clouds & Tropical Cyclones
Nov 06, 2025Abstract:Accurate forecasting of tropical cyclones (TCs) remains challenging due to limited satellite observations probing TC structure and difficulties in resolving cloud properties involved in TC intensification. Recent research has demonstrated the capabilities of machine learning methods for 3D cloud reconstruction from satellite observations. However, existing approaches have been restricted to regions where TCs are uncommon, and are poorly validated for intense storms. We introduce a new framework, based on a pre-training--fine-tuning pipeline, that learns from multiple satellites with global coverage to translate 2D satellite imagery into 3D cloud maps of relevant cloud properties. We apply our model to a custom-built TC dataset to evaluate performance in the most challenging and relevant conditions. We show that we can - for the first time - create global instantaneous 3D cloud maps and accurately reconstruct the 3D structure of intense storms. Our model not only extends available satellite observations but also provides estimates when observations are missing entirely. This is crucial for advancing our understanding of TC intensification and improving forecasts.
3D Cloud reconstruction through geospatially-aware Masked Autoencoders
Jan 03, 2025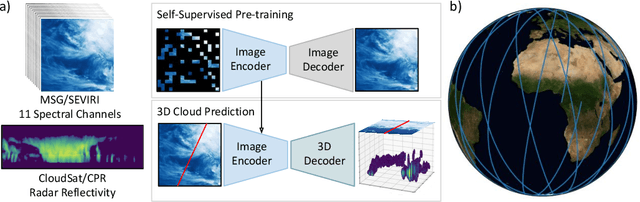
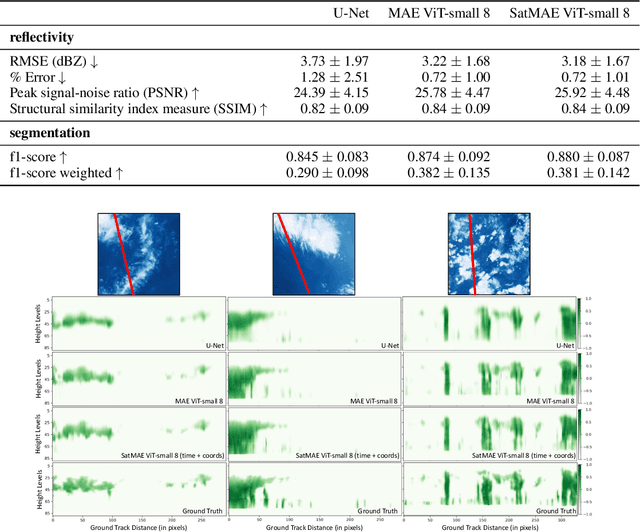
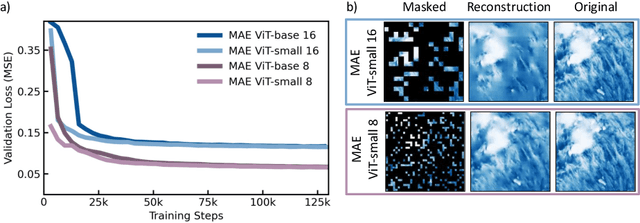
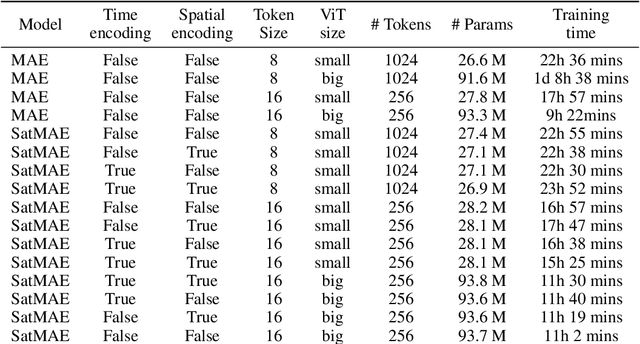
Abstract:Clouds play a key role in Earth's radiation balance with complex effects that introduce large uncertainties into climate models. Real-time 3D cloud data is essential for improving climate predictions. This study leverages geostationary imagery from MSG/SEVIRI and radar reflectivity measurements of cloud profiles from CloudSat/CPR to reconstruct 3D cloud structures. We first apply self-supervised learning (SSL) methods-Masked Autoencoders (MAE) and geospatially-aware SatMAE on unlabelled MSG images, and then fine-tune our models on matched image-profile pairs. Our approach outperforms state-of-the-art methods like U-Nets, and our geospatial encoding further improves prediction results, demonstrating the potential of SSL for cloud reconstruction.
A Comparative Study of Population-Graph Construction Methods and Graph Neural Networks for Brain Age Regression
Sep 26, 2023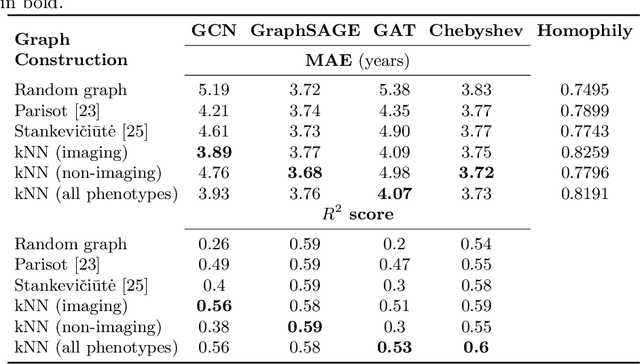
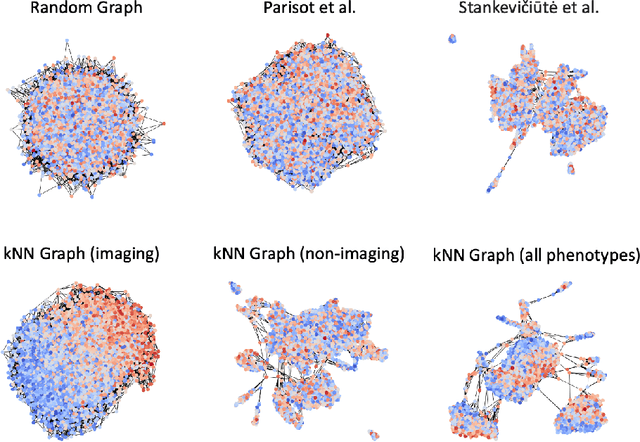
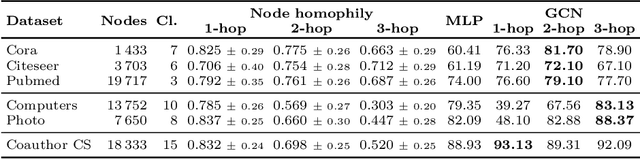
Abstract:The difference between the chronological and biological brain age of a subject can be an important biomarker for neurodegenerative diseases, thus brain age estimation can be crucial in clinical settings. One way to incorporate multimodal information into this estimation is through population graphs, which combine various types of imaging data and capture the associations among individuals within a population. In medical imaging, population graphs have demonstrated promising results, mostly for classification tasks. In most cases, the graph structure is pre-defined and remains static during training. However, extracting population graphs is a non-trivial task and can significantly impact the performance of Graph Neural Networks (GNNs), which are sensitive to the graph structure. In this work, we highlight the importance of a meaningful graph construction and experiment with different population-graph construction methods and their effect on GNN performance on brain age estimation. We use the homophily metric and graph visualizations to gain valuable quantitative and qualitative insights on the extracted graph structures. For the experimental evaluation, we leverage the UK Biobank dataset, which offers many imaging and non-imaging phenotypes. Our results indicate that architectures highly sensitive to the graph structure, such as Graph Convolutional Network (GCN) and Graph Attention Network (GAT), struggle with low homophily graphs, while other architectures, such as GraphSage and Chebyshev, are more robust across different homophily ratios. We conclude that static graph construction approaches are potentially insufficient for the task of brain age estimation and make recommendations for alternative research directions.
Multimodal brain age estimation using interpretable adaptive population-graph learning
Jul 19, 2023

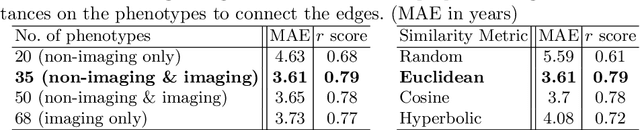

Abstract:Brain age estimation is clinically important as it can provide valuable information in the context of neurodegenerative diseases such as Alzheimer's. Population graphs, which include multimodal imaging information of the subjects along with the relationships among the population, have been used in literature along with Graph Convolutional Networks (GCNs) and have proved beneficial for a variety of medical imaging tasks. A population graph is usually static and constructed manually using non-imaging information. However, graph construction is not a trivial task and might significantly affect the performance of the GCN, which is inherently very sensitive to the graph structure. In this work, we propose a framework that learns a population graph structure optimized for the downstream task. An attention mechanism assigns weights to a set of imaging and non-imaging features (phenotypes), which are then used for edge extraction. The resulting graph is used to train the GCN. The entire pipeline can be trained end-to-end. Additionally, by visualizing the attention weights that were the most important for the graph construction, we increase the interpretability of the graph. We use the UK Biobank, which provides a large variety of neuroimaging and non-imaging phenotypes, to evaluate our method on brain age regression and classification. The proposed method outperforms competing static graph approaches and other state-of-the-art adaptive methods. We further show that the assigned attention scores indicate that there are both imaging and non-imaging phenotypes that are informative for brain age estimation and are in agreement with the relevant literature.
Extended Graph Assessment Metrics for Graph Neural Networks
Jul 13, 2023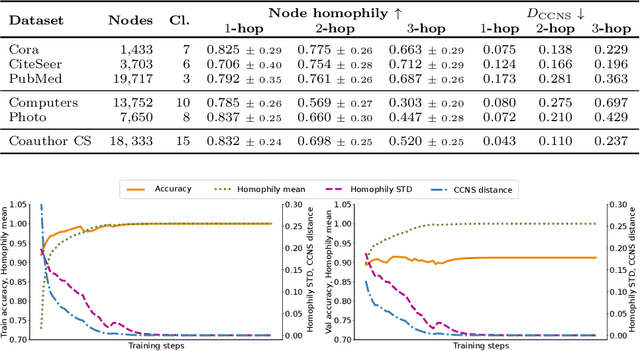
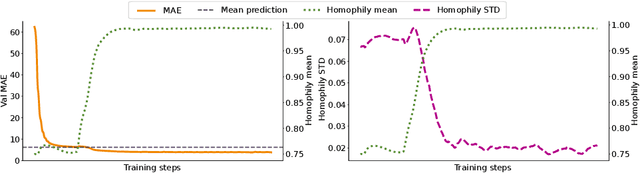
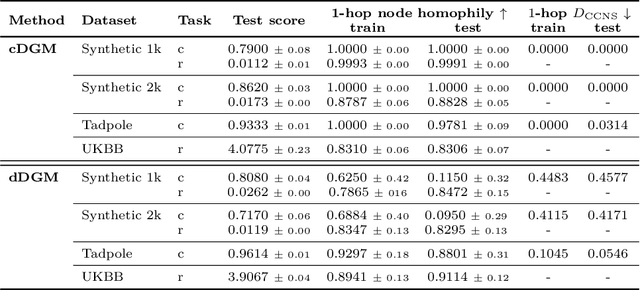
Abstract:When re-structuring patient cohorts into so-called population graphs, initially independent data points can be incorporated into one interconnected graph structure. This population graph can then be used for medical downstream tasks using graph neural networks (GNNs). The construction of a suitable graph structure is a challenging step in the learning pipeline that can have severe impact on model performance. To this end, different graph assessment metrics have been introduced to evaluate graph structures. However, these metrics are limited to classification tasks and discrete adjacency matrices, only covering a small subset of real-world applications. In this work, we introduce extended graph assessment metrics (GAMs) for regression tasks and continuous adjacency matrices. We focus on two GAMs in specific: \textit{homophily} and \textit{cross-class neighbourhood similarity} (CCNS). We extend the notion of GAMs to more than one hop, define homophily for regression tasks, as well as continuous adjacency matrices, and propose a light-weight CCNS distance for discrete and continuous adjacency matrices. We show the correlation of these metrics with model performance on different medical population graphs and under different learning settings.
GraphWalks: Efficient Shape Agnostic Geodesic Shortest Path Estimation
May 30, 2022



Abstract:Geodesic paths and distances are among the most popular intrinsic properties of 3D surfaces. Traditionally, geodesic paths on discrete polygon surfaces were computed using shortest path algorithms, such as Dijkstra. However, such algorithms have two major limitations. They are non-differentiable which limits their direct usage in learnable pipelines and they are considerably time demanding. To address such limitations and alleviate the computational burden, we propose a learnable network to approximate geodesic paths. The proposed method is comprised by three major components: a graph neural network that encodes node positions in a high dimensional space, a path embedding that describes previously visited nodes and a point classifier that selects the next point in the path. The proposed method provides efficient approximations of the shortest paths and geodesic distances estimations. Given that all of the components of our method are fully differentiable, it can be directly plugged into any learnable pipeline as well as customized under any differentiable constraint. We extensively evaluate the proposed method with several qualitative and quantitative experiments.
Voxel-level Importance Maps for Interpretable Brain Age Estimation
Aug 11, 2021

Abstract:Brain aging, and more specifically the difference between the chronological and the biological age of a person, may be a promising biomarker for identifying neurodegenerative diseases. For this purpose accurate prediction is important but the localisation of the areas that play a significant role in the prediction is also crucial, in order to gain clinicians' trust and reassurance about the performance of a prediction model. Most interpretability methods are focused on classification tasks and cannot be directly transferred to regression tasks. In this study, we focus on the task of brain age regression from 3D brain Magnetic Resonance (MR) images using a Convolutional Neural Network, termed prediction model. We interpret its predictions by extracting importance maps, which discover the parts of the brain that are the most important for brain age. In order to do so, we assume that voxels that are not useful for the regression are resilient to noise addition. We implement a noise model which aims to add as much noise as possible to the input without harming the performance of the prediction model. We average the importance maps of the subjects and end up with a population-based importance map, which displays the regions of the brain that are influential for the task. We test our method on 13,750 3D brain MR images from the UK Biobank, and our findings are consistent with the existing neuropathology literature, highlighting that the hippocampus and the ventricles are the most relevant regions for brain aging.
The Pitfalls of Sample Selection: A Case Study on Lung Nodule Classification
Aug 11, 2021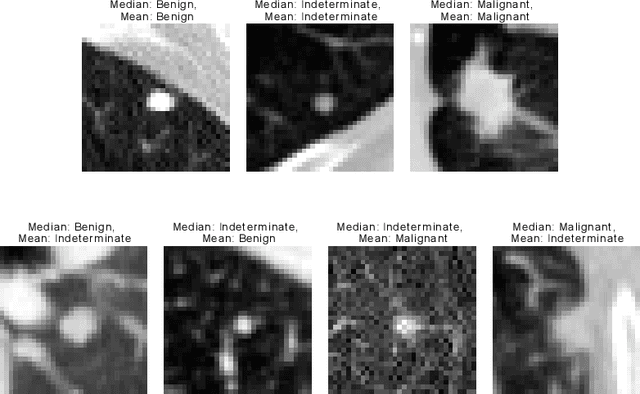
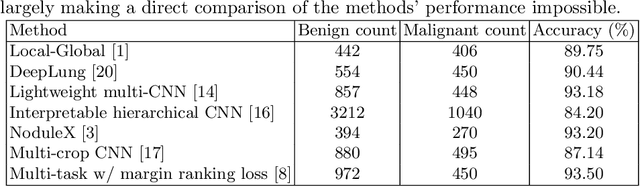
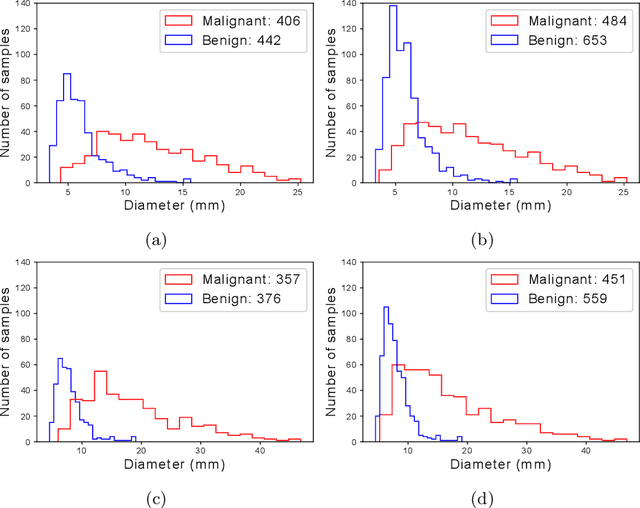
Abstract:Using publicly available data to determine the performance of methodological contributions is important as it facilitates reproducibility and allows scrutiny of the published results. In lung nodule classification, for example, many works report results on the publicly available LIDC dataset. In theory, this should allow a direct comparison of the performance of proposed methods and assess the impact of individual contributions. When analyzing seven recent works, however, we find that each employs a different data selection process, leading to largely varying total number of samples and ratios between benign and malignant cases. As each subset will have different characteristics with varying difficulty for classification, a direct comparison between the proposed methods is thus not always possible, nor fair. We study the particular effect of truthing when aggregating labels from multiple experts. We show that specific choices can have severe impact on the data distribution where it may be possible to achieve superior performance on one sample distribution but not on another. While we show that we can further improve on the state-of-the-art on one sample selection, we also find that on a more challenging sample selection, on the same database, the more advanced models underperform with respect to very simple baseline methods, highlighting that the selected data distribution may play an even more important role than the model architecture. This raises concerns about the validity of claimed methodological contributions. We believe the community should be aware of these pitfalls and make recommendations on how these can be avoided in future work.
The Effect of the Loss on Generalization: Empirical Study on Synthetic Lung Nodule Data
Aug 10, 2021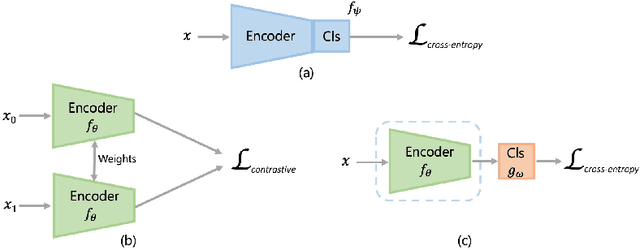
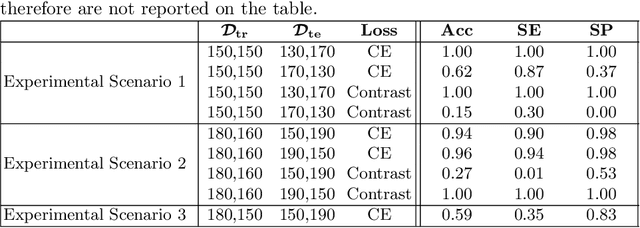
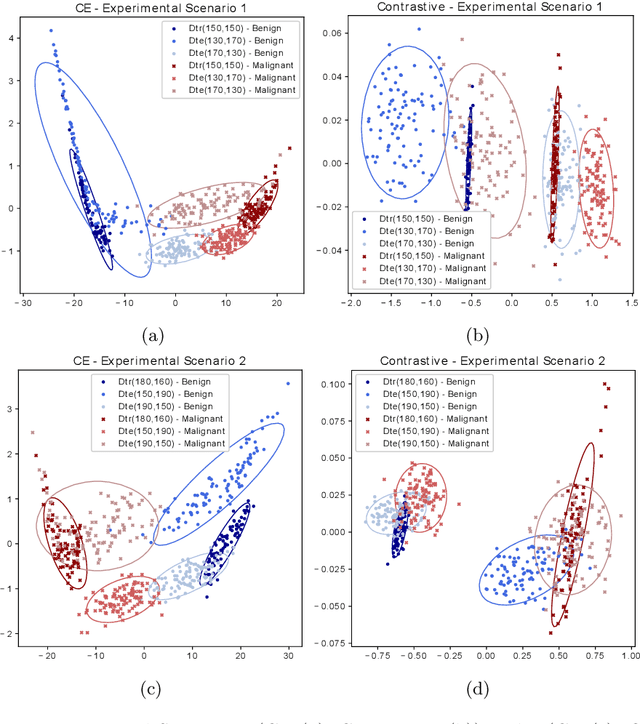
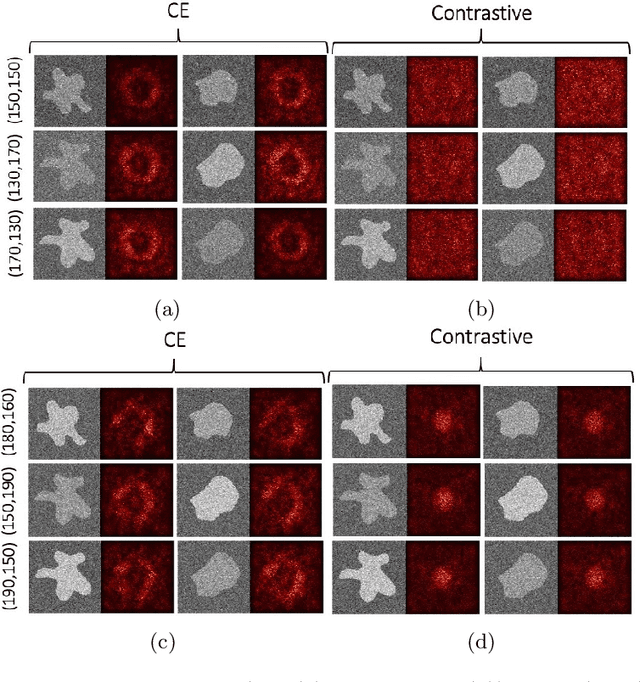
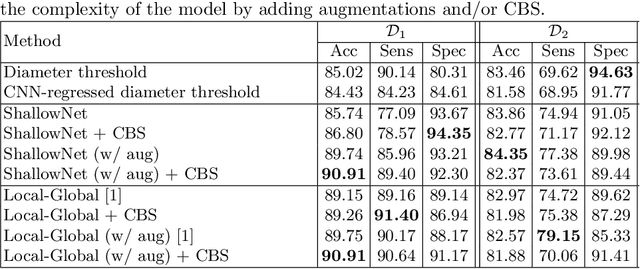
Abstract:Convolutional Neural Networks (CNNs) are widely used for image classification in a variety of fields, including medical imaging. While most studies deploy cross-entropy as the loss function in such tasks, a growing number of approaches have turned to a family of contrastive learning-based losses. Even though performance metrics such as accuracy, sensitivity and specificity are regularly used for the evaluation of CNN classifiers, the features that these classifiers actually learn are rarely identified and their effect on the classification performance on out-of-distribution test samples is insufficiently explored. In this paper, motivated by the real-world task of lung nodule classification, we investigate the features that a CNN learns when trained and tested on different distributions of a synthetic dataset with controlled modes of variation. We show that different loss functions lead to different features being learned and consequently affect the generalization ability of the classifier on unseen data. This study provides some important insights into the design of deep learning solutions for medical imaging tasks.
Patch-based Brain Age Estimation from MR Images
Oct 01, 2020
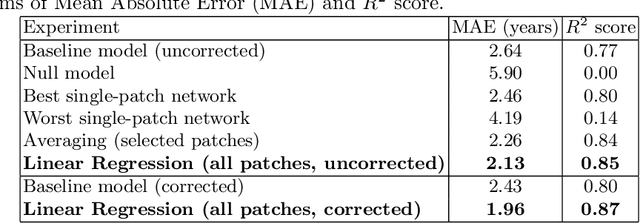


Abstract:Brain age estimation from Magnetic Resonance Images (MRI) derives the difference between a subject's biological brain age and their chronological age. This is a potential biomarker for neurodegeneration, e.g. as part of Alzheimer's disease. Early detection of neurodegeneration manifesting as a higher brain age can potentially facilitate better medical care and planning for affected individuals. Many studies have been proposed for the prediction of chronological age from brain MRI using machine learning and specifically deep learning techniques. Contrary to most studies, which use the whole brain volume, in this study, we develop a new deep learning approach that uses 3D patches of the brain as well as convolutional neural networks (CNNs) to develop a localised brain age estimator. In this way, we can obtain a visualization of the regions that play the most important role for estimating brain age, leading to more anatomically driven and interpretable results, and thus confirming relevant literature which suggests that the ventricles and the hippocampus are the areas that are most informative. In addition, we leverage this knowledge in order to improve the overall performance on the task of age estimation by combining the results of different patches using an ensemble method, such as averaging or linear regression. The network is trained on the UK Biobank dataset and the method achieves state-of-the-art results with a Mean Absolute Error of 2.46 years for purely regional estimates, and 2.13 years for an ensemble of patches before bias correction, while 1.96 years after bias correction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge