Alexander Hammers
School of Biomedical Engineering and Imaging Sciences, King's College London, U.K
Localising Shortcut Learning in Pixel Space via Ordinal Scoring Correlations for Attribution Representations (OSCAR)
Dec 21, 2025



Abstract:Deep neural networks often exploit shortcuts. These are spurious cues which are associated with output labels in the training data but are unrelated to task semantics. When the shortcut features are associated with sensitive attributes, shortcut learning can lead to biased model performance. Existing methods for localising and understanding shortcut learning are mostly based upon qualitative, image-level inspection and assume cues are human-visible, limiting their use in domains such as medical imaging. We introduce OSCAR (Ordinal Scoring Correlations for Attribution Representations), a model-agnostic framework for quantifying shortcut learning and localising shortcut features. OSCAR converts image-level task attribution maps into dataset-level rank profiles of image regions and compares them across three models: a balanced baseline model (BA), a test model (TS), and a sensitive attribute predictor (SA). By computing pairwise, partial, and deviation-based correlations on these rank profiles, we produce a set of quantitative metrics that characterise the degree of shortcut reliance for TS, together with a ranking of image-level regions that contribute most to it. Experiments on CelebA, CheXpert, and ADNI show that our correlations are (i) stable across seeds and partitions, (ii) sensitive to the level of association between shortcut features and output labels in the training data, and (iii) able to distinguish localised from diffuse shortcut features. As an illustration of the utility of our method, we show how worst-group performance disparities can be reduced using a simple test-time attenuation approach based on the identified shortcut regions. OSCAR provides a lightweight, pixel-space audit that yields statistical decision rules and spatial maps, enabling users to test, localise, and mitigate shortcut reliance. The code is available at https://github.com/acharaakshit/oscar
Invisible Attributes, Visible Biases: Exploring Demographic Shortcuts in MRI-based Alzheimer's Disease Classification
Sep 11, 2025Abstract:Magnetic resonance imaging (MRI) is the gold standard for brain imaging. Deep learning (DL) algorithms have been proposed to aid in the diagnosis of diseases such as Alzheimer's disease (AD) from MRI scans. However, DL algorithms can suffer from shortcut learning, in which spurious features, not directly related to the output label, are used for prediction. When these features are related to protected attributes, they can lead to performance bias against underrepresented protected groups, such as those defined by race and sex. In this work, we explore the potential for shortcut learning and demographic bias in DL based AD diagnosis from MRI. We first investigate if DL algorithms can identify race or sex from 3D brain MRI scans to establish the presence or otherwise of race and sex based distributional shifts. Next, we investigate whether training set imbalance by race or sex can cause a drop in model performance, indicating shortcut learning and bias. Finally, we conduct a quantitative and qualitative analysis of feature attributions in different brain regions for both the protected attribute and AD classification tasks. Through these experiments, and using multiple datasets and DL models (ResNet and SwinTransformer), we demonstrate the existence of both race and sex based shortcut learning and bias in DL based AD classification. Our work lays the foundation for fairer DL diagnostic tools in brain MRI. The code is provided at https://github.com/acharaakshit/ShortMR
Systematic Review of Pituitary Gland and Pituitary Adenoma Automatic Segmentation Techniques in Magnetic Resonance Imaging
Jun 24, 2025Abstract:Purpose: Accurate segmentation of both the pituitary gland and adenomas from magnetic resonance imaging (MRI) is essential for diagnosis and treatment of pituitary adenomas. This systematic review evaluates automatic segmentation methods for improving the accuracy and efficiency of MRI-based segmentation of pituitary adenomas and the gland itself. Methods: We reviewed 34 studies that employed automatic and semi-automatic segmentation methods. We extracted and synthesized data on segmentation techniques and performance metrics (such as Dice overlap scores). Results: The majority of reviewed studies utilized deep learning approaches, with U-Net-based models being the most prevalent. Automatic methods yielded Dice scores of 0.19--89.00\% for pituitary gland and 4.60--96.41\% for adenoma segmentation. Semi-automatic methods reported 80.00--92.10\% for pituitary gland and 75.90--88.36\% for adenoma segmentation. Conclusion: Most studies did not report important metrics such as MR field strength, age and adenoma size. Automated segmentation techniques such as U-Net-based models show promise, especially for adenoma segmentation, but further improvements are needed to achieve consistently good performance in small structures like the normal pituitary gland. Continued innovation and larger, diverse datasets are likely critical to enhancing clinical applicability.
Personalized MR-Informed Diffusion Models for 3D PET Image Reconstruction
Jun 04, 2025Abstract:Recent work has shown improved lesion detectability and flexibility to reconstruction hyperparameters (e.g. scanner geometry or dose level) when PET images are reconstructed by leveraging pre-trained diffusion models. Such methods train a diffusion model (without sinogram data) on high-quality, but still noisy, PET images. In this work, we propose a simple method for generating subject-specific PET images from a dataset of multi-subject PET-MR scans, synthesizing "pseudo-PET" images by transforming between different patients' anatomy using image registration. The images we synthesize retain information from the subject's MR scan, leading to higher resolution and the retention of anatomical features compared to the original set of PET images. With simulated and real [$^{18}$F]FDG datasets, we show that pre-training a personalized diffusion model with subject-specific "pseudo-PET" images improves reconstruction accuracy with low-count data. In particular, the method shows promise in combining information from a guidance MR scan without overly imposing anatomical features, demonstrating an improved trade-off between reconstructing PET-unique image features versus features present in both PET and MR. We believe this approach for generating and utilizing synthetic data has further applications to medical imaging tasks, particularly because patient-specific PET images can be generated without resorting to generative deep learning or large training datasets.
Likelihood-Scheduled Score-Based Generative Modeling for Fully 3D PET Image Reconstruction
Dec 05, 2024Abstract:Medical image reconstruction with pre-trained score-based generative models (SGMs) has advantages over other existing state-of-the-art deep-learned reconstruction methods, including improved resilience to different scanner setups and advanced image distribution modeling. SGM-based reconstruction has recently been applied to simulated positron emission tomography (PET) datasets, showing improved contrast recovery for out-of-distribution lesions relative to the state-of-the-art. However, existing methods for SGM-based reconstruction from PET data suffer from slow reconstruction, burdensome hyperparameter tuning and slice inconsistency effects (in 3D). In this work, we propose a practical methodology for fully 3D reconstruction that accelerates reconstruction and reduces the number of critical hyperparameters by matching the likelihood of an SGM's reverse diffusion process to a current iterate of the maximum-likelihood expectation maximization algorithm. Using the example of low-count reconstruction from simulated $[^{18}$F]DPA-714 datasets, we show our methodology can match or improve on the NRMSE and SSIM of existing state-of-the-art SGM-based PET reconstruction while reducing reconstruction time and the need for hyperparameter tuning. We evaluate our methodology against state-of-the-art supervised and conventional reconstruction algorithms. Finally, we demonstrate a first-ever implementation of SGM-based reconstruction for real 3D PET data, specifically $[^{18}$F]DPA-714 data, where we integrate perpendicular pre-trained SGMs to eliminate slice inconsistency issues.
Multi-Subject Image Synthesis as a Generative Prior for Single-Subject PET Image Reconstruction
Dec 05, 2024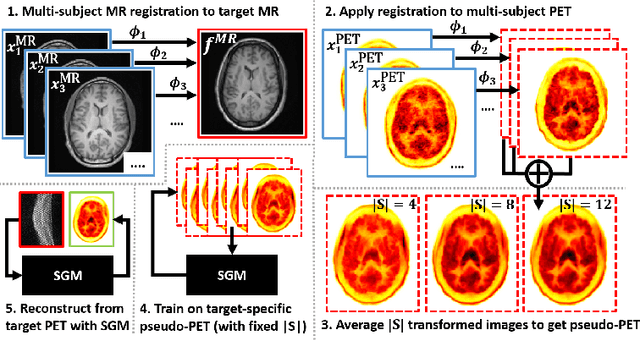
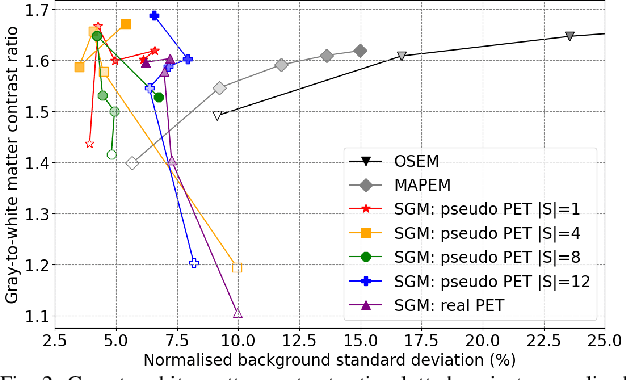
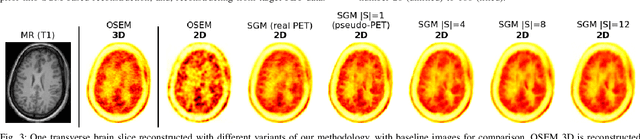
Abstract:Large high-quality medical image datasets are difficult to acquire but necessary for many deep learning applications. For positron emission tomography (PET), reconstructed image quality is limited by inherent Poisson noise. We propose a novel method for synthesising diverse and realistic pseudo-PET images with improved signal-to-noise ratio. We also show how our pseudo-PET images may be exploited as a generative prior for single-subject PET image reconstruction. Firstly, we perform deep-learned deformable registration of multi-subject magnetic resonance (MR) images paired to multi-subject PET images. We then use the anatomically-learned deformation fields to transform multiple PET images to the same reference space, before averaging random subsets of the transformed multi-subject data to form a large number of varying pseudo-PET images. We observe that using MR information for registration imbues the resulting pseudo-PET images with improved anatomical detail compared to the originals. We consider applications to PET image reconstruction, by generating pseudo-PET images in the same space as the intended single-subject reconstruction and using them as training data for a diffusion model-based reconstruction method. We show visual improvement and reduced background noise in our 2D reconstructions as compared to OSEM, MAP-EM and an existing state-of-the-art diffusion model-based approach. Our method shows the potential for utilising highly subject-specific prior information within a generative reconstruction framework. Future work may compare the benefits of our approach to explicitly MR-guided reconstruction methodologies.
Generative-Model-Based Fully 3D PET Image Reconstruction by Conditional Diffusion Sampling
Dec 05, 2024Abstract:Score-based generative models (SGMs) have recently shown promising results for image reconstruction on simulated positron emission tomography (PET) datasets. In this work we have developed and implemented practical methodology for 3D image reconstruction with SGMs, and perform (to our knowledge) the first SGM-based reconstruction of real fully 3D PET data. We train an SGM on full-count reference brain images, and extend methodology to allow SGM-based reconstructions at very low counts (1% of original, to simulate low-dose or short-duration scanning). We then perform reconstructions for multiple independent realisations of 1% count data, allowing us to analyse the bias and variance characteristics of the method. We sample from the learned posterior distribution of the generative algorithm to calculate uncertainty images for our reconstructions. We evaluate the method's performance on real full- and low-count PET data and compare with conventional OSEM and MAP-EM baselines, showing that our SGM-based low-count reconstructions match full-dose reconstructions more closely and in a bias-variance trade-off comparison, our SGM-reconstructed images have lower variance than existing baselines. Future work will compare to supervised deep-learned methods, with other avenues for investigation including how data conditioning affects the SGM's posterior distribution and the algorithm's performance with different tracers.
A Comparative Study of Population-Graph Construction Methods and Graph Neural Networks for Brain Age Regression
Sep 26, 2023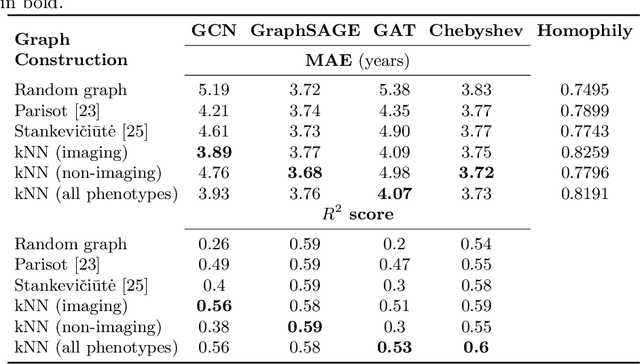
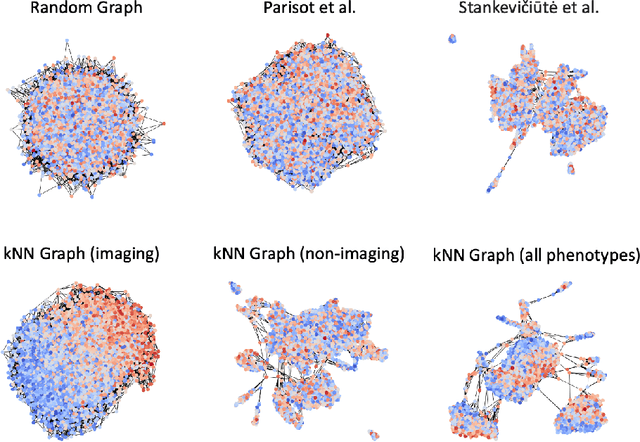
Abstract:The difference between the chronological and biological brain age of a subject can be an important biomarker for neurodegenerative diseases, thus brain age estimation can be crucial in clinical settings. One way to incorporate multimodal information into this estimation is through population graphs, which combine various types of imaging data and capture the associations among individuals within a population. In medical imaging, population graphs have demonstrated promising results, mostly for classification tasks. In most cases, the graph structure is pre-defined and remains static during training. However, extracting population graphs is a non-trivial task and can significantly impact the performance of Graph Neural Networks (GNNs), which are sensitive to the graph structure. In this work, we highlight the importance of a meaningful graph construction and experiment with different population-graph construction methods and their effect on GNN performance on brain age estimation. We use the homophily metric and graph visualizations to gain valuable quantitative and qualitative insights on the extracted graph structures. For the experimental evaluation, we leverage the UK Biobank dataset, which offers many imaging and non-imaging phenotypes. Our results indicate that architectures highly sensitive to the graph structure, such as Graph Convolutional Network (GCN) and Graph Attention Network (GAT), struggle with low homophily graphs, while other architectures, such as GraphSage and Chebyshev, are more robust across different homophily ratios. We conclude that static graph construction approaches are potentially insufficient for the task of brain age estimation and make recommendations for alternative research directions.
Multimodal brain age estimation using interpretable adaptive population-graph learning
Jul 19, 2023

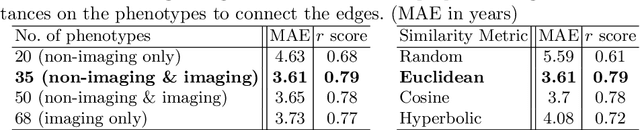

Abstract:Brain age estimation is clinically important as it can provide valuable information in the context of neurodegenerative diseases such as Alzheimer's. Population graphs, which include multimodal imaging information of the subjects along with the relationships among the population, have been used in literature along with Graph Convolutional Networks (GCNs) and have proved beneficial for a variety of medical imaging tasks. A population graph is usually static and constructed manually using non-imaging information. However, graph construction is not a trivial task and might significantly affect the performance of the GCN, which is inherently very sensitive to the graph structure. In this work, we propose a framework that learns a population graph structure optimized for the downstream task. An attention mechanism assigns weights to a set of imaging and non-imaging features (phenotypes), which are then used for edge extraction. The resulting graph is used to train the GCN. The entire pipeline can be trained end-to-end. Additionally, by visualizing the attention weights that were the most important for the graph construction, we increase the interpretability of the graph. We use the UK Biobank, which provides a large variety of neuroimaging and non-imaging phenotypes, to evaluate our method on brain age regression and classification. The proposed method outperforms competing static graph approaches and other state-of-the-art adaptive methods. We further show that the assigned attention scores indicate that there are both imaging and non-imaging phenotypes that are informative for brain age estimation and are in agreement with the relevant literature.
A Study of Demographic Bias in CNN-based Brain MR Segmentation
Aug 13, 2022



Abstract:Convolutional neural networks (CNNs) are increasingly being used to automate the segmentation of brain structures in magnetic resonance (MR) images for research studies. In other applications, CNN models have been shown to exhibit bias against certain demographic groups when they are under-represented in the training sets. In this work, we investigate whether CNN models for brain MR segmentation have the potential to contain sex or race bias when trained with imbalanced training sets. We train multiple instances of the FastSurferCNN model using different levels of sex imbalance in white subjects. We evaluate the performance of these models separately for white male and white female test sets to assess sex bias, and furthermore evaluate them on black male and black female test sets to assess potential racial bias. We find significant sex and race bias effects in segmentation model performance. The biases have a strong spatial component, with some brain regions exhibiting much stronger bias than others. Overall, our results suggest that race bias is more significant than sex bias. Our study demonstrates the importance of considering race and sex balance when forming training sets for CNN-based brain MR segmentation, to avoid maintaining or even exacerbating existing health inequalities through biased research study findings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge