Oliver D. Howes
Multi-Subject Image Synthesis as a Generative Prior for Single-Subject PET Image Reconstruction
Dec 05, 2024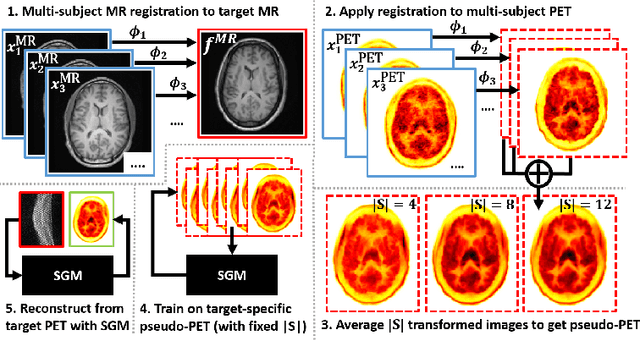
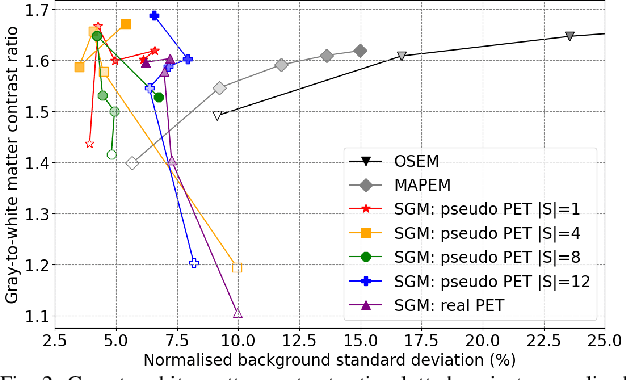
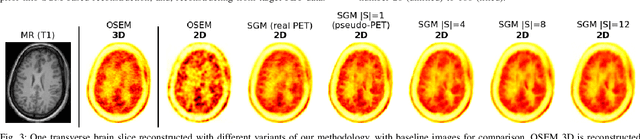
Abstract:Large high-quality medical image datasets are difficult to acquire but necessary for many deep learning applications. For positron emission tomography (PET), reconstructed image quality is limited by inherent Poisson noise. We propose a novel method for synthesising diverse and realistic pseudo-PET images with improved signal-to-noise ratio. We also show how our pseudo-PET images may be exploited as a generative prior for single-subject PET image reconstruction. Firstly, we perform deep-learned deformable registration of multi-subject magnetic resonance (MR) images paired to multi-subject PET images. We then use the anatomically-learned deformation fields to transform multiple PET images to the same reference space, before averaging random subsets of the transformed multi-subject data to form a large number of varying pseudo-PET images. We observe that using MR information for registration imbues the resulting pseudo-PET images with improved anatomical detail compared to the originals. We consider applications to PET image reconstruction, by generating pseudo-PET images in the same space as the intended single-subject reconstruction and using them as training data for a diffusion model-based reconstruction method. We show visual improvement and reduced background noise in our 2D reconstructions as compared to OSEM, MAP-EM and an existing state-of-the-art diffusion model-based approach. Our method shows the potential for utilising highly subject-specific prior information within a generative reconstruction framework. Future work may compare the benefits of our approach to explicitly MR-guided reconstruction methodologies.
Likelihood-Scheduled Score-Based Generative Modeling for Fully 3D PET Image Reconstruction
Dec 05, 2024Abstract:Medical image reconstruction with pre-trained score-based generative models (SGMs) has advantages over other existing state-of-the-art deep-learned reconstruction methods, including improved resilience to different scanner setups and advanced image distribution modeling. SGM-based reconstruction has recently been applied to simulated positron emission tomography (PET) datasets, showing improved contrast recovery for out-of-distribution lesions relative to the state-of-the-art. However, existing methods for SGM-based reconstruction from PET data suffer from slow reconstruction, burdensome hyperparameter tuning and slice inconsistency effects (in 3D). In this work, we propose a practical methodology for fully 3D reconstruction that accelerates reconstruction and reduces the number of critical hyperparameters by matching the likelihood of an SGM's reverse diffusion process to a current iterate of the maximum-likelihood expectation maximization algorithm. Using the example of low-count reconstruction from simulated $[^{18}$F]DPA-714 datasets, we show our methodology can match or improve on the NRMSE and SSIM of existing state-of-the-art SGM-based PET reconstruction while reducing reconstruction time and the need for hyperparameter tuning. We evaluate our methodology against state-of-the-art supervised and conventional reconstruction algorithms. Finally, we demonstrate a first-ever implementation of SGM-based reconstruction for real 3D PET data, specifically $[^{18}$F]DPA-714 data, where we integrate perpendicular pre-trained SGMs to eliminate slice inconsistency issues.
Generative-Model-Based Fully 3D PET Image Reconstruction by Conditional Diffusion Sampling
Dec 05, 2024Abstract:Score-based generative models (SGMs) have recently shown promising results for image reconstruction on simulated positron emission tomography (PET) datasets. In this work we have developed and implemented practical methodology for 3D image reconstruction with SGMs, and perform (to our knowledge) the first SGM-based reconstruction of real fully 3D PET data. We train an SGM on full-count reference brain images, and extend methodology to allow SGM-based reconstructions at very low counts (1% of original, to simulate low-dose or short-duration scanning). We then perform reconstructions for multiple independent realisations of 1% count data, allowing us to analyse the bias and variance characteristics of the method. We sample from the learned posterior distribution of the generative algorithm to calculate uncertainty images for our reconstructions. We evaluate the method's performance on real full- and low-count PET data and compare with conventional OSEM and MAP-EM baselines, showing that our SGM-based low-count reconstructions match full-dose reconstructions more closely and in a bias-variance trade-off comparison, our SGM-reconstructed images have lower variance than existing baselines. Future work will compare to supervised deep-learned methods, with other avenues for investigation including how data conditioning affects the SGM's posterior distribution and the algorithm's performance with different tracers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge