Huaqi Qiu
Parametric shape models for vessels learned from segmentations via differentiable voxelization
Jul 03, 2025Abstract:Vessels are complex structures in the body that have been studied extensively in multiple representations. While voxelization is the most common of them, meshes and parametric models are critical in various applications due to their desirable properties. However, these representations are typically extracted through segmentations and used disjointly from each other. We propose a framework that joins the three representations under differentiable transformations. By leveraging differentiable voxelization, we automatically extract a parametric shape model of the vessels through shape-to-segmentation fitting, where we learn shape parameters from segmentations without the explicit need for ground-truth shape parameters. The vessel is parametrized as centerlines and radii using cubic B-splines, ensuring smoothness and continuity by construction. Meshes are differentiably extracted from the learned shape parameters, resulting in high-fidelity meshes that can be manipulated post-fit. Our method can accurately capture the geometry of complex vessels, as demonstrated by the volumetric fits in experiments on aortas, aneurysms, and brain vessels.
Evaluation of Alignment-Regularity Characteristics in Deformable Image Registration
Mar 10, 2025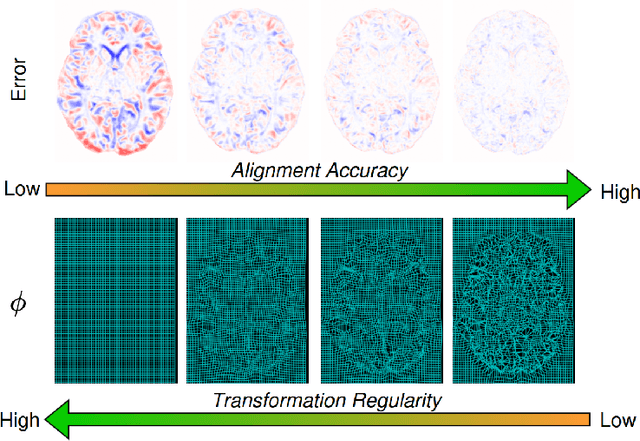
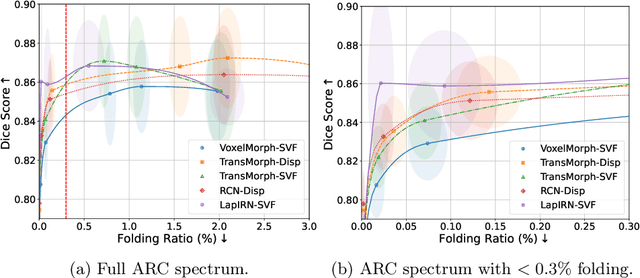
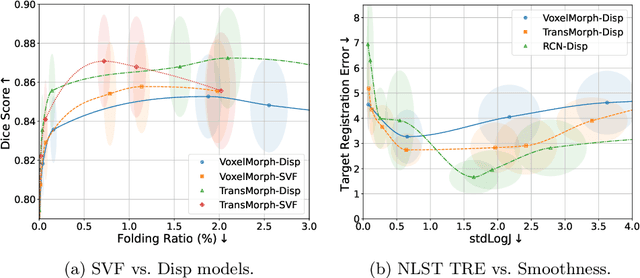
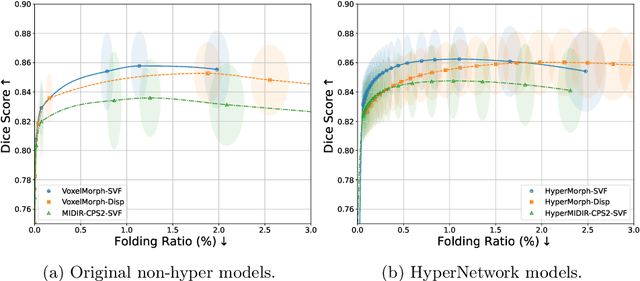
Abstract:Evaluating deformable image registration (DIR) is challenging due to the inherent trade-off between achieving high alignment accuracy and maintaining deformation regularity. In this work, we introduce a novel evaluation scheme based on the alignment-regularity characteristic (ARC) to systematically capture and analyze this trade-off. We first introduce the ARC curves, which describe the performance of a given registration algorithm as a spectrum measured by alignment and regularity metrics. We further adopt a HyperNetwork-based approach that learns to continuously interpolate across the full regularization range, accelerating the construction and improving the sample density of ARC curves. We empirically demonstrate our evaluation scheme using representative learning-based deformable image registration methods with various network architectures and transformation models on two public datasets. We present a range of findings not evident from existing evaluation practices and provide general recommendations for model evaluation and selection using our evaluation scheme. All code relevant is made publicly available.
Image registration is a geometric deep learning task
Dec 17, 2024



Abstract:Data-driven deformable image registration methods predominantly rely on operations that process grid-like inputs. However, applying deformable transformations to an image results in a warped space that deviates from a rigid grid structure. Consequently, data-driven approaches with sequential deformations have to apply grid resampling operations between each deformation step. While artifacts caused by resampling are negligible in high-resolution images, the resampling of sparse, high-dimensional feature grids introduces errors that affect the deformation modeling process. Taking inspiration from Lagrangian reference frames of deformation fields, our work introduces a novel paradigm for data-driven deformable image registration that utilizes geometric deep-learning principles to model deformations without grid requirements. Specifically, we model image features as a set of nodes that freely move in Euclidean space, update their coordinates under graph operations, and dynamically readjust their local neighborhoods. We employ this formulation to construct a multi-resolution deformable registration model, where deformation layers iteratively refine the overall transformation at each resolution without intermediate resampling operations on the feature grids. We investigate our method's ability to fully deformably capture large deformations across a number of medical imaging registration tasks. In particular, we apply our approach (GeoReg) to the registration of inter-subject brain MR images and inhale-exhale lung CT images, showing on par performance with the current state-of-the-art methods. We believe our contribution open up avenues of research to reduce the black-box nature of current learned registration paradigms by explicitly modeling the transformation within the architecture.
MAD: Modality Agnostic Distance Measure for Image Registration
Sep 06, 2023Abstract:Multi-modal image registration is a crucial pre-processing step in many medical applications. However, it is a challenging task due to the complex intensity relationships between different imaging modalities, which can result in large discrepancy in image appearance. The success of multi-modal image registration, whether it is conventional or learning based, is predicated upon the choice of an appropriate distance (or similarity) measure. Particularly, deep learning registration algorithms lack in accuracy or even fail completely when attempting to register data from an "unseen" modality. In this work, we present Modality Agnostic Distance (MAD), a deep image distance}] measure that utilises random convolutions to learn the inherent geometry of the images while being robust to large appearance changes. Random convolutions are geometry-preserving modules which we use to simulate an infinite number of synthetic modalities alleviating the need for aligned paired data during training. We can therefore train MAD on a mono-modal dataset and successfully apply it to a multi-modal dataset. We demonstrate that not only can MAD affinely register multi-modal images successfully, but it has also a larger capture range than traditional measures such as Mutual Information and Normalised Gradient Fields.
Conditional Deformable Image Registration with Spatially-Variant and Adaptive Regularization
Mar 19, 2023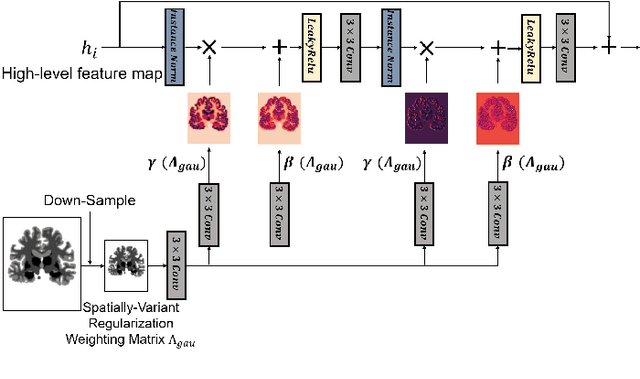
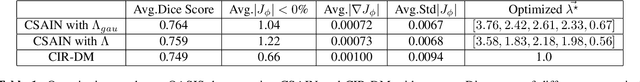
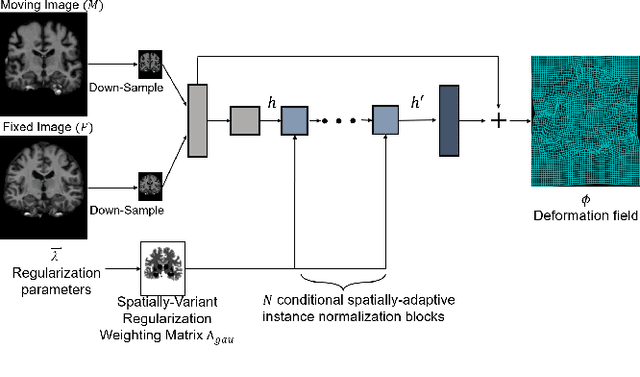
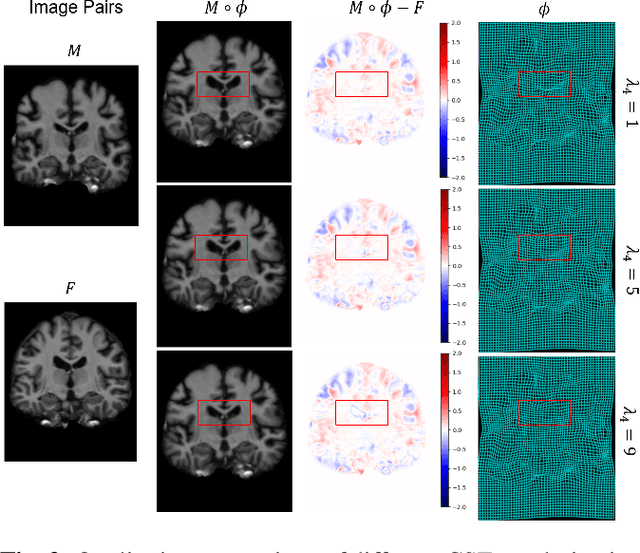
Abstract:Deep learning-based image registration approaches have shown competitive performance and run-time advantages compared to conventional image registration methods. However, existing learning-based approaches mostly require to train separate models with respect to different regularization hyperparameters for manual hyperparameter searching and often do not allow spatially-variant regularization. In this work, we propose a learning-based registration approach based on a novel conditional spatially adaptive instance normalization (CSAIN) to address these challenges. The proposed method introduces a spatially-variant regularization and learns its effect of achieving spatially-adaptive regularization by conditioning the registration network on the hyperparameter matrix via CSAIN. This allows varying of spatially adaptive regularization at inference to obtain multiple plausible deformations with a single pre-trained model. Additionally, the proposed method enables automatic hyperparameter optimization to avoid manual hyperparameter searching. Experiments show that our proposed method outperforms the baseline approaches while achieving spatially-variant and adaptive regularization.
CHeart: A Conditional Spatio-Temporal Generative Model for Cardiac Anatomy
Jan 30, 2023Abstract:Two key questions in cardiac image analysis are to assess the anatomy and motion of the heart from images; and to understand how they are associated with non-imaging clinical factors such as gender, age and diseases. While the first question can often be addressed by image segmentation and motion tracking algorithms, our capability to model and to answer the second question is still limited. In this work, we propose a novel conditional generative model to describe the 4D spatio-temporal anatomy of the heart and its interaction with non-imaging clinical factors. The clinical factors are integrated as the conditions of the generative modelling, which allows us to investigate how these factors influence the cardiac anatomy. We evaluate the model performance in mainly two tasks, anatomical sequence completion and sequence generation. The model achieves a high performance in anatomical sequence completion, comparable to or outperforming other state-of-the-art generative models. In terms of sequence generation, given clinical conditions, the model can generate realistic synthetic 4D sequential anatomies that share similar distributions with the real data.
Generative Modelling of the Ageing Heart with Cross-Sectional Imaging and Clinical Data
Aug 28, 2022



Abstract:Cardiovascular disease, the leading cause of death globally, is an age-related disease. Understanding the morphological and functional changes of the heart during ageing is a key scientific question, the answer to which will help us define important risk factors of cardiovascular disease and monitor disease progression. In this work, we propose a novel conditional generative model to describe the changes of 3D anatomy of the heart during ageing. The proposed model is flexible and allows integration of multiple clinical factors (e.g. age, gender) into the generating process. We train the model on a large-scale cross-sectional dataset of cardiac anatomies and evaluate on both cross-sectional and longitudinal datasets. The model demonstrates excellent performance in predicting the longitudinal evolution of the ageing heart and modelling its data distribution.
Learn2Reg: comprehensive multi-task medical image registration challenge, dataset and evaluation in the era of deep learning
Dec 23, 2021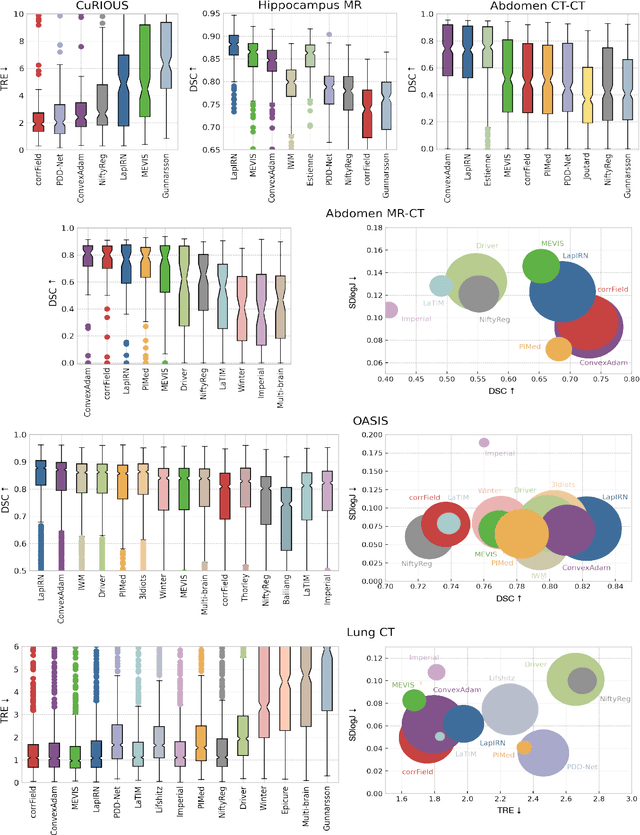
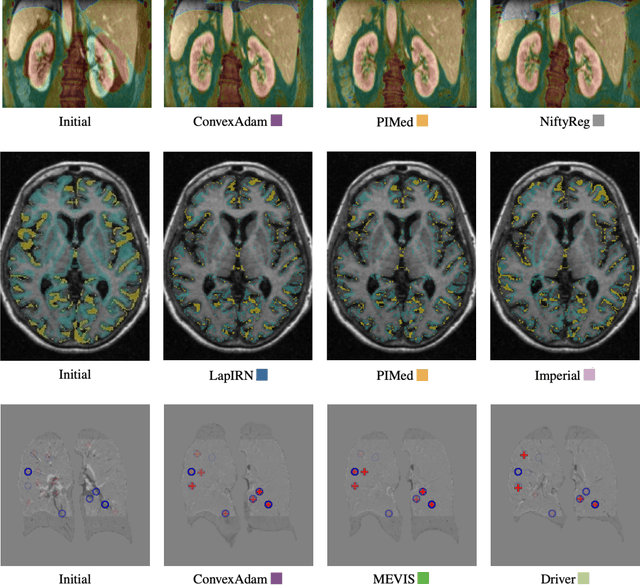
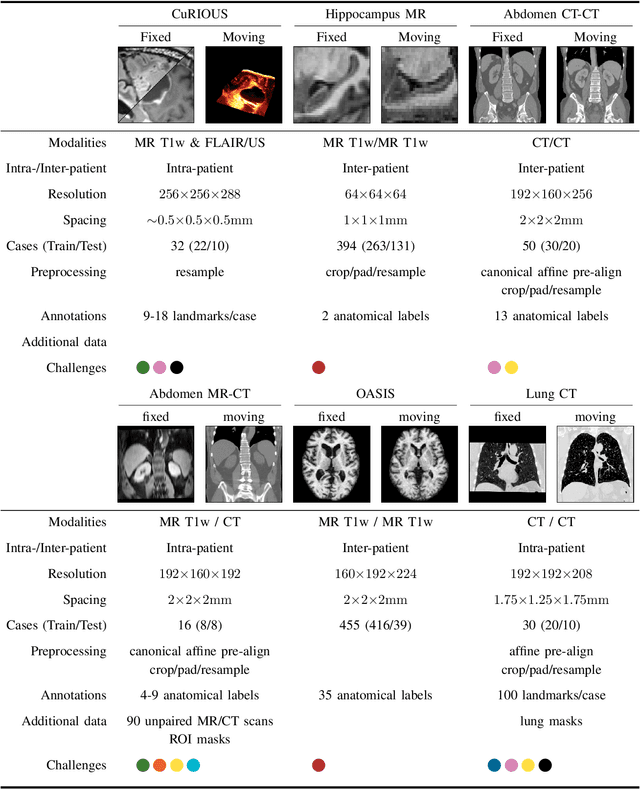
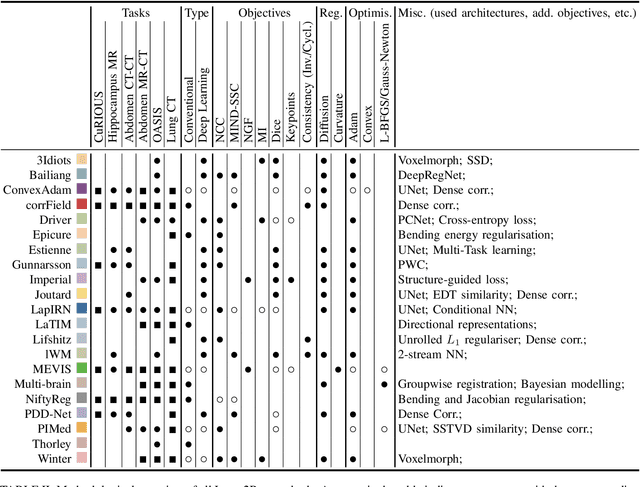
Abstract:Image registration is a fundamental medical image analysis task, and a wide variety of approaches have been proposed. However, only a few studies have comprehensively compared medical image registration approaches on a wide range of clinically relevant tasks, in part because of the lack of availability of such diverse data. This limits the development of registration methods, the adoption of research advances into practice, and a fair benchmark across competing approaches. The Learn2Reg challenge addresses these limitations by providing a multi-task medical image registration benchmark for comprehensive characterisation of deformable registration algorithms. A continuous evaluation will be possible at https://learn2reg.grand-challenge.org. Learn2Reg covers a wide range of anatomies (brain, abdomen, and thorax), modalities (ultrasound, CT, MR), availability of annotations, as well as intra- and inter-patient registration evaluation. We established an easily accessible framework for training and validation of 3D registration methods, which enabled the compilation of results of over 65 individual method submissions from more than 20 unique teams. We used a complementary set of metrics, including robustness, accuracy, plausibility, and runtime, enabling unique insight into the current state-of-the-art of medical image registration. This paper describes datasets, tasks, evaluation methods and results of the challenge, and the results of further analysis of transferability to new datasets, the importance of label supervision, and resulting bias.
GraDIRN: Learning Iterative Gradient Descent-based Energy Minimization for Deformable Image Registration
Dec 07, 2021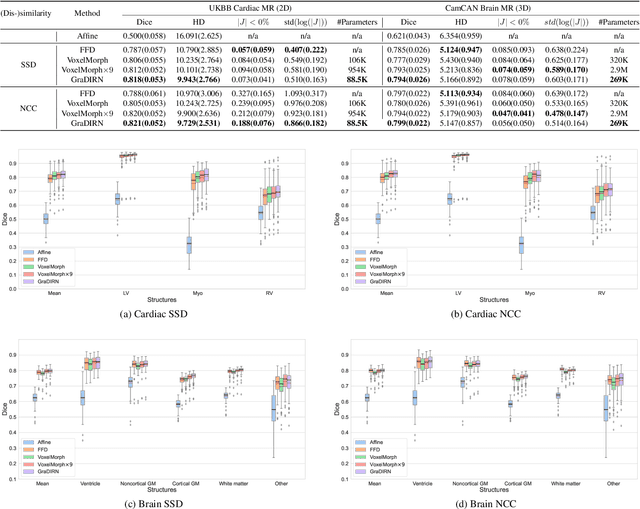

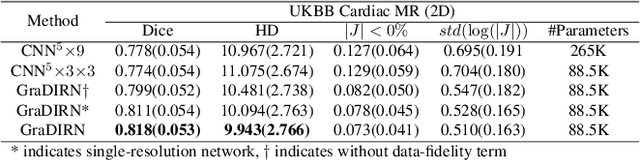

Abstract:We present a Gradient Descent-based Image Registration Network (GraDIRN) for learning deformable image registration by embedding gradient-based iterative energy minimization in a deep learning framework. Traditional image registration algorithms typically use iterative energy-minimization optimization to find the optimal transformation between a pair of images, which is time-consuming when many iterations are needed. In contrast, recent learning-based methods amortize this costly iterative optimization by training deep neural networks so that registration of one pair of images can be achieved by fast network forward pass after training. Motivated by successes in image reconstruction techniques that combine deep learning with the mathematical structure of iterative variational energy optimization, we formulate a novel registration network based on multi-resolution gradient descent energy minimization. The forward pass of the network takes explicit image dissimilarity gradient steps and generalized regularization steps parameterized by Convolutional Neural Networks (CNN) for a fixed number of iterations. We use auto-differentiation to derive the forward computational graph for the explicit image dissimilarity gradient w.r.t. the transformation, so arbitrary image dissimilarity metrics and transformation models can be used without complex and error-prone gradient derivations. We demonstrate that this approach achieves state-of-the-art registration performance while using fewer learnable parameters through extensive evaluations on registration tasks using 2D cardiac MR images and 3D brain MR images.
Uncertainty quantification in non-rigid image registration via stochastic gradient Markov chain Monte Carlo
Oct 25, 2021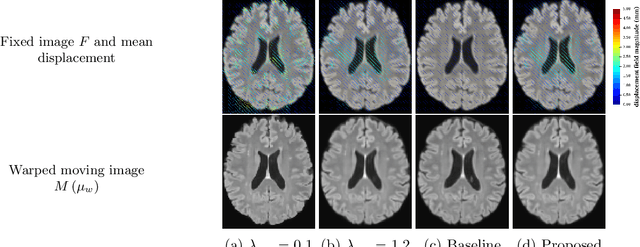
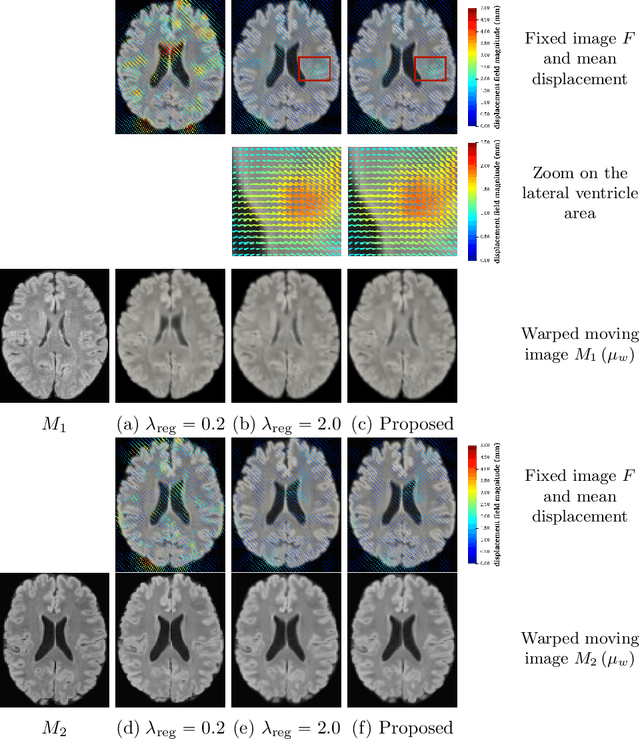

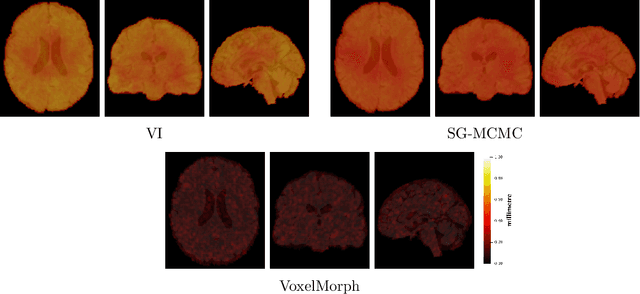
Abstract:We develop a new Bayesian model for non-rigid registration of three-dimensional medical images, with a focus on uncertainty quantification. Probabilistic registration of large images with calibrated uncertainty estimates is difficult for both computational and modelling reasons. To address the computational issues, we explore connections between the Markov chain Monte Carlo by backpropagation and the variational inference by backpropagation frameworks, in order to efficiently draw samples from the posterior distribution of transformation parameters. To address the modelling issues, we formulate a Bayesian model for image registration that overcomes the existing barriers when using a dense, high-dimensional, and diffeomorphic transformation parametrisation. This results in improved calibration of uncertainty estimates. We compare the model in terms of both image registration accuracy and uncertainty quantification to VoxelMorph, a state-of-the-art image registration model based on deep learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge