Nil Stolt-Ansó
Fast and Explicit: Slice-to-Volume Reconstruction via 3D Gaussian Primitives with Analytic Point Spread Function Modeling
Dec 16, 2025Abstract:Recovering high-fidelity 3D images from sparse or degraded 2D images is a fundamental challenge in medical imaging, with broad applications ranging from 3D ultrasound reconstruction to MRI super-resolution. In the context of fetal MRI, high-resolution 3D reconstruction of the brain from motion-corrupted low-resolution 2D acquisitions is a prerequisite for accurate neurodevelopmental diagnosis. While implicit neural representations (INRs) have recently established state-of-the-art performance in self-supervised slice-to-volume reconstruction (SVR), they suffer from a critical computational bottleneck: accurately modeling the image acquisition physics requires expensive stochastic Monte Carlo sampling to approximate the point spread function (PSF). In this work, we propose a shift from neural network based implicit representations to Gaussian based explicit representations. By parameterizing the HR 3D image volume as a field of anisotropic Gaussian primitives, we leverage the property of Gaussians being closed under convolution and thus derive a \textit{closed-form analytical solution} for the forward model. This formulation reduces the previously intractable acquisition integral to an exact covariance addition ($\mathbfΣ_{obs} = \mathbfΣ_{HR} + \mathbfΣ_{PSF}$), effectively bypassing the need for compute-intensive stochastic sampling while ensuring exact gradient propagation. We demonstrate that our approach matches the reconstruction quality of self-supervised state-of-the-art SVR frameworks while delivering a 5$\times$--10$\times$ speed-up on neonatal and fetal data. With convergence often reached in under 30 seconds, our framework paves the way towards translation into clinical routine of real-time fetal 3D MRI. Code will be public at {https://github.com/m-dannecker/Gaussian-Primitives-for-Fast-SVR}.
Reconstruction-free segmentation from undersampled k-space using transformers
Nov 05, 2025Abstract:Motivation: High acceleration factors place a limit on MRI image reconstruction. This limit is extended to segmentation models when treating these as subsequent independent processes. Goal: Our goal is to produce segmentations directly from sparse k-space measurements without the need for intermediate image reconstruction. Approach: We employ a transformer architecture to encode global k-space information into latent features. The produced latent vectors condition queried coordinates during decoding to generate segmentation class probabilities. Results: The model is able to produce better segmentations across high acceleration factors than image-based segmentation baselines. Impact: Cardiac segmentation directly from undersampled k-space samples circumvents the need for an intermediate image reconstruction step. This allows the potential to assess myocardial structure and function on higher acceleration factors than methods that rely on images as input.
Image registration is a geometric deep learning task
Dec 17, 2024



Abstract:Data-driven deformable image registration methods predominantly rely on operations that process grid-like inputs. However, applying deformable transformations to an image results in a warped space that deviates from a rigid grid structure. Consequently, data-driven approaches with sequential deformations have to apply grid resampling operations between each deformation step. While artifacts caused by resampling are negligible in high-resolution images, the resampling of sparse, high-dimensional feature grids introduces errors that affect the deformation modeling process. Taking inspiration from Lagrangian reference frames of deformation fields, our work introduces a novel paradigm for data-driven deformable image registration that utilizes geometric deep-learning principles to model deformations without grid requirements. Specifically, we model image features as a set of nodes that freely move in Euclidean space, update their coordinates under graph operations, and dynamically readjust their local neighborhoods. We employ this formulation to construct a multi-resolution deformable registration model, where deformation layers iteratively refine the overall transformation at each resolution without intermediate resampling operations on the feature grids. We investigate our method's ability to fully deformably capture large deformations across a number of medical imaging registration tasks. In particular, we apply our approach (GeoReg) to the registration of inter-subject brain MR images and inhale-exhale lung CT images, showing on par performance with the current state-of-the-art methods. We believe our contribution open up avenues of research to reduce the black-box nature of current learned registration paradigms by explicitly modeling the transformation within the architecture.
Direct Cardiac Segmentation from Undersampled K-space Using Transformers
May 31, 2024


Abstract:The prevailing deep learning-based methods of predicting cardiac segmentation involve reconstructed magnetic resonance (MR) images. The heavy dependency of segmentation approaches on image quality significantly limits the acceleration rate in fast MR reconstruction. Moreover, the practice of treating reconstruction and segmentation as separate sequential processes leads to artifact generation and information loss in the intermediate stage. These issues pose a great risk to achieving high-quality outcomes. To leverage the redundant k-space information overlooked in this dual-step pipeline, we introduce a novel approach to directly deriving segmentations from sparse k-space samples using a transformer (DiSK). DiSK operates by globally extracting latent features from 2D+time k-space data with attention blocks and subsequently predicting the segmentation label of query points. We evaluate our model under various acceleration factors (ranging from 4 to 64) and compare against two image-based segmentation baselines. Our model consistently outperforms the baselines in Dice and Hausdorff distances across foreground classes for all presented sampling rates.
Intensity-based 3D motion correction for cardiac MR images
Mar 31, 2024



Abstract:Cardiac magnetic resonance (CMR) image acquisition requires subjects to hold their breath while 2D cine images are acquired. This process assumes that the heart remains in the same position across all slices. However, differences in breathhold positions or patient motion introduce 3D slice misalignments. In this work, we propose an algorithm that simultaneously aligns all SA and LA slices by maximizing the pair-wise intensity agreement between their intersections. Unlike previous works, our approach is formulated as a subject-specific optimization problem and requires no prior knowledge of the underlying anatomy. We quantitatively demonstrate that the proposed method is robust against a large range of rotations and translations by synthetically misaligning 10 motion-free datasets and aligning them back using the proposed method.
NISF: Neural Implicit Segmentation Functions
Sep 15, 2023Abstract:Segmentation of anatomical shapes from medical images has taken an important role in the automation of clinical measurements. While typical deep-learning segmentation approaches are performed on discrete voxels, the underlying objects being analysed exist in a real-valued continuous space. Approaches that rely on convolutional neural networks (CNNs) are limited to grid-like inputs and not easily applicable to sparse or partial measurements. We propose a novel family of image segmentation models that tackle many of CNNs' shortcomings: Neural Implicit Segmentation Functions (NISF). Our framework takes inspiration from the field of neural implicit functions where a network learns a mapping from a real-valued coordinate-space to a shape representation. NISFs have the ability to segment anatomical shapes in high-dimensional continuous spaces. Training is not limited to voxelized grids, and covers applications with sparse and partial data. Interpolation between observations is learnt naturally in the training procedure and requires no post-processing. Furthermore, NISFs allow the leveraging of learnt shape priors to make predictions for regions outside of the original image plane. We go on to show the framework achieves dice scores of 0.87 $\pm$ 0.045 on a (3D+t) short-axis cardiac segmentation task using the UK Biobank dataset. We also provide a qualitative analysis on our frameworks ability to perform segmentation and image interpolation on unseen regions of an image volume at arbitrary resolutions.
Global k-Space Interpolation for Dynamic MRI Reconstruction using Masked Image Modeling
Jul 24, 2023


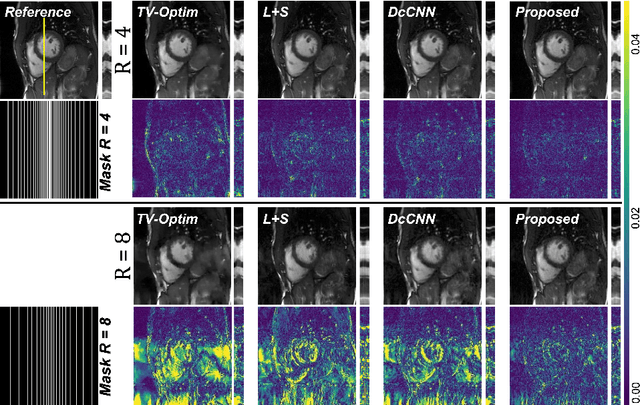
Abstract:In dynamic Magnetic Resonance Imaging (MRI), k-space is typically undersampled due to limited scan time, resulting in aliasing artifacts in the image domain. Hence, dynamic MR reconstruction requires not only modeling spatial frequency components in the x and y directions of k-space but also considering temporal redundancy. Most previous works rely on image-domain regularizers (priors) to conduct MR reconstruction. In contrast, we focus on interpolating the undersampled k-space before obtaining images with Fourier transform. In this work, we connect masked image modeling with k-space interpolation and propose a novel Transformer-based k-space Global Interpolation Network, termed k-GIN. Our k-GIN learns global dependencies among low- and high-frequency components of 2D+t k-space and uses it to interpolate unsampled data. Further, we propose a novel k-space Iterative Refinement Module (k-IRM) to enhance the high-frequency components learning. We evaluate our approach on 92 in-house 2D+t cardiac MR subjects and compare it to MR reconstruction methods with image-domain regularizers. Experiments show that our proposed k-space interpolation method quantitatively and qualitatively outperforms baseline methods. Importantly, the proposed approach achieves substantially higher robustness and generalizability in cases of highly-undersampled MR data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge