Kerstin Hammernik
Towards a Unified Theoretical Framework for Self-Supervised MRI Reconstruction
Jan 08, 2026Abstract:The demand for high-resolution, non-invasive imaging continues to drive innovation in magnetic resonance imaging (MRI), yet prolonged acquisition times hinder accessibility and real-time applications. While deep learning-based reconstruction methods have accelerated MRI, their predominant supervised paradigm depends on fully-sampled reference data that are challenging to acquire. Recently, self-supervised learning (SSL) approaches have emerged as promising alternatives, but most are empirically designed and fragmented. Therefore, we introduce UNITS (Unified Theory for Self-supervision), a general framework for self-supervised MRI reconstruction. UNITS unifies prior SSL strategies within a common formalism, enabling consistent interpretation and systematic benchmarking. We prove that SSL can achieve the same expected performance as supervised learning. Under this theoretical guarantee, we introduce sampling stochasticity and flexible data utilization, which improve network generalization under out-of-domain distributions and stabilize training. Together, these contributions establish UNITS as a theoretical foundation and a practical paradigm for interpretable, generalizable, and clinically applicable self-supervised MRI reconstruction.
Reconstruction-free segmentation from undersampled k-space using transformers
Nov 05, 2025Abstract:Motivation: High acceleration factors place a limit on MRI image reconstruction. This limit is extended to segmentation models when treating these as subsequent independent processes. Goal: Our goal is to produce segmentations directly from sparse k-space measurements without the need for intermediate image reconstruction. Approach: We employ a transformer architecture to encode global k-space information into latent features. The produced latent vectors condition queried coordinates during decoding to generate segmentation class probabilities. Results: The model is able to produce better segmentations across high acceleration factors than image-based segmentation baselines. Impact: Cardiac segmentation directly from undersampled k-space samples circumvents the need for an intermediate image reconstruction step. This allows the potential to assess myocardial structure and function on higher acceleration factors than methods that rely on images as input.
Self-supervised feature learning for cardiac Cine MR image reconstruction
May 29, 2025Abstract:We propose a self-supervised feature learning assisted reconstruction (SSFL-Recon) framework for MRI reconstruction to address the limitation of existing supervised learning methods. Although recent deep learning-based methods have shown promising performance in MRI reconstruction, most require fully-sampled images for supervised learning, which is challenging in practice considering long acquisition times under respiratory or organ motion. Moreover, nearly all fully-sampled datasets are obtained from conventional reconstruction of mildly accelerated datasets, thus potentially biasing the achievable performance. The numerous undersampled datasets with different accelerations in clinical practice, hence, remain underutilized. To address these issues, we first train a self-supervised feature extractor on undersampled images to learn sampling-insensitive features. The pre-learned features are subsequently embedded in the self-supervised reconstruction network to assist in removing artifacts. Experiments were conducted retrospectively on an in-house 2D cardiac Cine dataset, including 91 cardiovascular patients and 38 healthy subjects. The results demonstrate that the proposed SSFL-Recon framework outperforms existing self-supervised MRI reconstruction methods and even exhibits comparable or better performance to supervised learning up to $16\times$ retrospective undersampling. The feature learning strategy can effectively extract global representations, which have proven beneficial in removing artifacts and increasing generalization ability during reconstruction.
Motion-Robust T2* Quantification from Gradient Echo MRI with Physics-Informed Deep Learning
Feb 24, 2025Abstract:Purpose: T2* quantification from gradient echo magnetic resonance imaging is particularly affected by subject motion due to the high sensitivity to magnetic field inhomogeneities, which are influenced by motion and might cause signal loss. Thus, motion correction is crucial to obtain high-quality T2* maps. Methods: We extend our previously introduced learning-based physics-informed motion correction method, PHIMO, by utilizing acquisition knowledge to enhance the reconstruction performance for challenging motion patterns and increase PHIMO's robustness to varying strengths of magnetic field inhomogeneities across the brain. We perform comprehensive evaluations regarding motion detection accuracy and image quality for data with simulated and real motion. Results: Our extended version of PHIMO outperforms the learning-based baseline methods both qualitatively and quantitatively with respect to line detection and image quality. Moreover, PHIMO performs on-par with a conventional state-of-the-art motion correction method for T2* quantification from gradient echo MRI, which relies on redundant data acquisition. Conclusion: PHIMO's competitive motion correction performance, combined with a reduction in acquisition time by over 40% compared to the state-of-the-art method, make it a promising solution for motion-robust T2* quantification in research settings and clinical routine.
PISCO: Self-Supervised k-Space Regularization for Improved Neural Implicit k-Space Representations of Dynamic MRI
Jan 16, 2025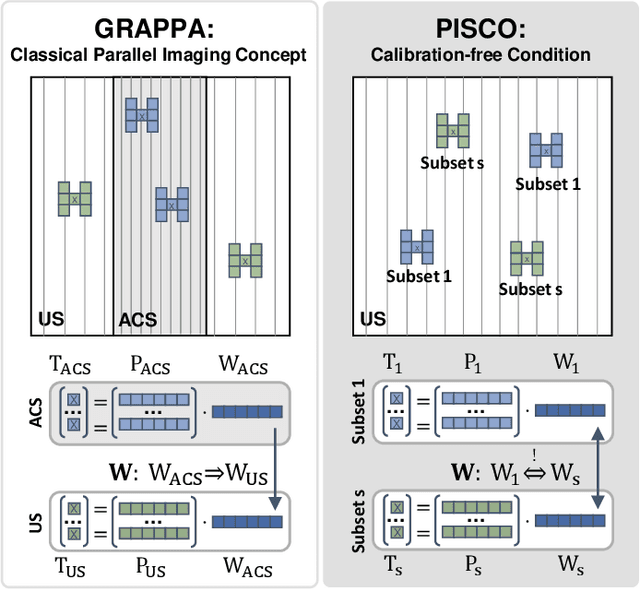
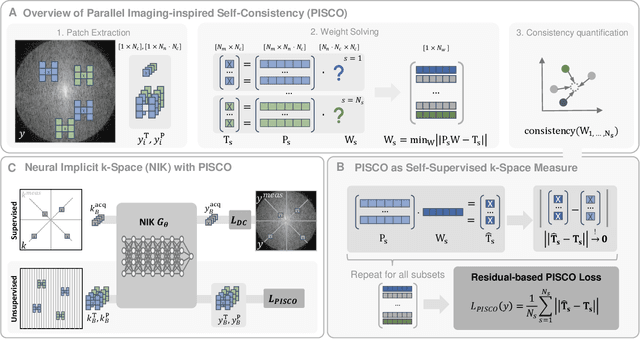
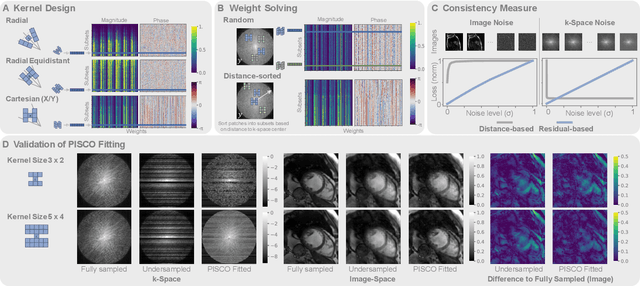
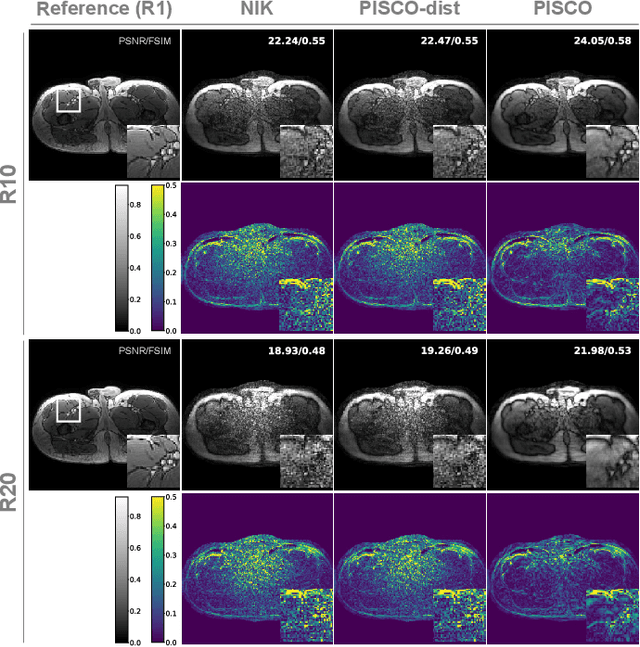
Abstract:Neural implicit k-space representations (NIK) have shown promising results for dynamic magnetic resonance imaging (MRI) at high temporal resolutions. Yet, reducing acquisition time, and thereby available training data, results in severe performance drops due to overfitting. To address this, we introduce a novel self-supervised k-space loss function $\mathcal{L}_\mathrm{PISCO}$, applicable for regularization of NIK-based reconstructions. The proposed loss function is based on the concept of parallel imaging-inspired self-consistency (PISCO), enforcing a consistent global k-space neighborhood relationship without requiring additional data. Quantitative and qualitative evaluations on static and dynamic MR reconstructions show that integrating PISCO significantly improves NIK representations. Particularly for high acceleration factors (R$\geq$54), NIK with PISCO achieves superior spatio-temporal reconstruction quality compared to state-of-the-art methods. Furthermore, an extensive analysis of the loss assumptions and stability shows PISCO's potential as versatile self-supervised k-space loss function for further applications and architectures. Code is available at: https://github.com/compai-lab/2025-pisco-spieker
Subspace Implicit Neural Representations for Real-Time Cardiac Cine MR Imaging
Dec 17, 2024
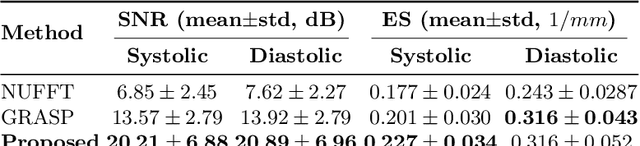
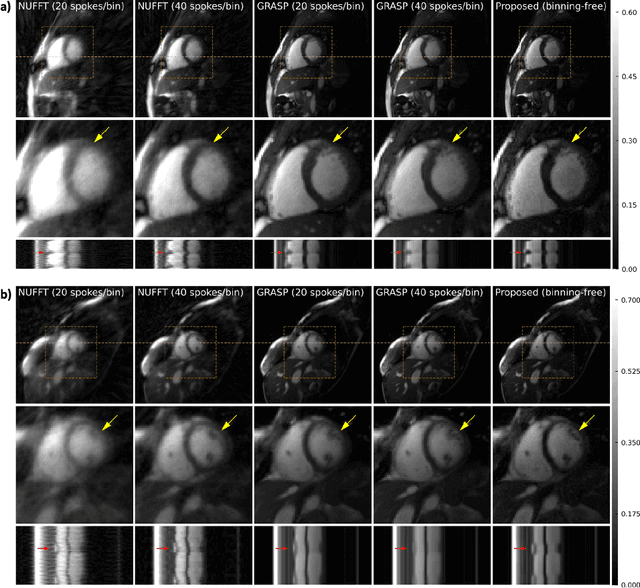
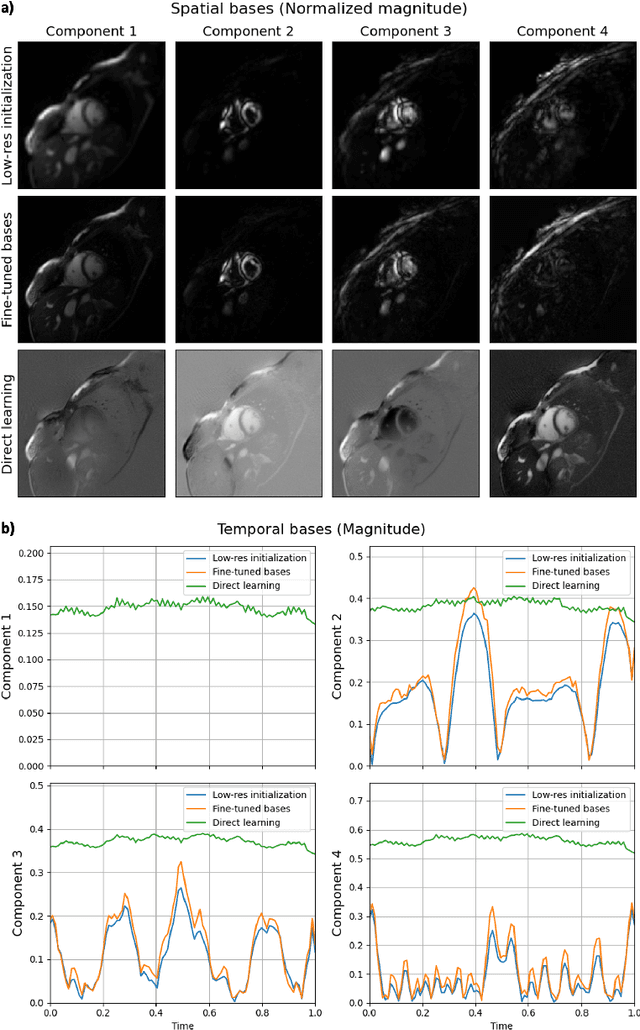
Abstract:Conventional cardiac cine MRI methods rely on retrospective gating, which limits temporal resolution and the ability to capture continuous cardiac dynamics, particularly in patients with arrhythmias and beat-to-beat variations. To address these challenges, we propose a reconstruction framework based on subspace implicit neural representations for real-time cardiac cine MRI of continuously sampled radial data. This approach employs two multilayer perceptrons to learn spatial and temporal subspace bases, leveraging the low-rank properties of cardiac cine MRI. Initialized with low-resolution reconstructions, the networks are fine-tuned using spoke-specific loss functions to recover spatial details and temporal fidelity. Our method directly utilizes the continuously sampled radial k-space spokes during training, thereby eliminating the need for binning and non-uniform FFT. This approach achieves superior spatial and temporal image quality compared to conventional binned methods at the acceleration rate of 10 and 20, demonstrating potential for high-resolution imaging of dynamic cardiac events and enhancing diagnostic capability.
Highly efficient non-rigid registration in k-space with application to cardiac Magnetic Resonance Imaging
Oct 24, 2024



Abstract:In Magnetic Resonance Imaging (MRI), high temporal-resolved motion can be useful for image acquisition and reconstruction, MR-guided radiotherapy, dynamic contrast-enhancement, flow and perfusion imaging, and functional assessment of motion patterns in cardiovascular, abdominal, peristaltic, fetal, or musculoskeletal imaging. Conventionally, these motion estimates are derived through image-based registration, a particularly challenging task for complex motion patterns and high dynamic resolution. The accelerated scans in such applications result in imaging artifacts that compromise the motion estimation. In this work, we propose a novel self-supervised deep learning-based framework, dubbed the Local-All Pass Attention Network (LAPANet), for non-rigid motion estimation directly from the acquired accelerated Fourier space, i.e. k-space. The proposed approach models non-rigid motion as the cumulative sum of local translational displacements, following the Local All-Pass (LAP) registration technique. LAPANet was evaluated on cardiac motion estimation across various sampling trajectories and acceleration rates. Our results demonstrate superior accuracy compared to prior conventional and deep learning-based registration methods, accommodating as few as 2 lines/frame in a Cartesian trajectory and 3 spokes/frame in a non-Cartesian trajectory. The achieved high temporal resolution (less than 5 ms) for non-rigid motion opens new avenues for motion detection, tracking and correction in dynamic and real-time MRI applications.
Attention Incorporated Network for Sharing Low-rank, Image and K-space Information during MR Image Reconstruction to Achieve Single Breath-hold Cardiac Cine Imaging
Jul 03, 2024Abstract:Cardiac Cine Magnetic Resonance Imaging (MRI) provides an accurate assessment of heart morphology and function in clinical practice. However, MRI requires long acquisition times, with recent deep learning-based methods showing great promise to accelerate imaging and enhance reconstruction quality. Existing networks exhibit some common limitations that constrain further acceleration possibilities, including single-domain learning, reliance on a single regularization term, and equal feature contribution. To address these limitations, we propose to embed information from multiple domains, including low-rank, image, and k-space, in a novel deep learning network for MRI reconstruction, which we denote as A-LIKNet. A-LIKNet adopts a parallel-branch structure, enabling independent learning in the k-space and image domain. Coupled information sharing layers realize the information exchange between domains. Furthermore, we introduce attention mechanisms into the network to assign greater weights to more critical coils or important temporal frames. Training and testing were conducted on an in-house dataset, including 91 cardiovascular patients and 38 healthy subjects scanned with 2D cardiac Cine using retrospective undersampling. Additionally, we evaluated A-LIKNet on the real-time 8x prospectively undersampled data from the OCMR dataset. The results demonstrate that our proposed A-LIKNet outperforms existing methods and provides high-quality reconstructions. The network can effectively reconstruct highly retrospectively undersampled dynamic MR images up to 24x accelerations, indicating its potential for single breath-hold imaging.
Direct Cardiac Segmentation from Undersampled K-space Using Transformers
May 31, 2024


Abstract:The prevailing deep learning-based methods of predicting cardiac segmentation involve reconstructed magnetic resonance (MR) images. The heavy dependency of segmentation approaches on image quality significantly limits the acceleration rate in fast MR reconstruction. Moreover, the practice of treating reconstruction and segmentation as separate sequential processes leads to artifact generation and information loss in the intermediate stage. These issues pose a great risk to achieving high-quality outcomes. To leverage the redundant k-space information overlooked in this dual-step pipeline, we introduce a novel approach to directly deriving segmentations from sparse k-space samples using a transformer (DiSK). DiSK operates by globally extracting latent features from 2D+time k-space data with attention blocks and subsequently predicting the segmentation label of query points. We evaluate our model under various acceleration factors (ranging from 4 to 64) and compare against two image-based segmentation baselines. Our model consistently outperforms the baselines in Dice and Hausdorff distances across foreground classes for all presented sampling rates.
Attention-aware non-rigid image registration for accelerated MR imaging
Apr 26, 2024Abstract:Accurate motion estimation at high acceleration factors enables rapid motion-compensated reconstruction in Magnetic Resonance Imaging (MRI) without compromising the diagnostic image quality. In this work, we introduce an attention-aware deep learning-based framework that can perform non-rigid pairwise registration for fully sampled and accelerated MRI. We extract local visual representations to build similarity maps between the registered image pairs at multiple resolution levels and additionally leverage long-range contextual information using a transformer-based module to alleviate ambiguities in the presence of artifacts caused by undersampling. We combine local and global dependencies to perform simultaneous coarse and fine motion estimation. The proposed method was evaluated on in-house acquired fully sampled and accelerated data of 101 patients and 62 healthy subjects undergoing cardiac and thoracic MRI. The impact of motion estimation accuracy on the downstream task of motion-compensated reconstruction was analyzed. We demonstrate that our model derives reliable and consistent motion fields across different sampling trajectories (Cartesian and radial) and acceleration factors of up to 16x for cardiac motion and 30x for respiratory motion and achieves superior image quality in motion-compensated reconstruction qualitatively and quantitatively compared to conventional and recent deep learning-based approaches. The code is publicly available at https://github.com/lab-midas/GMARAFT.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge