Wenqi Huang
on behalf of the PREDICTOM consortium
Reconstruction-free segmentation from undersampled k-space using transformers
Nov 05, 2025Abstract:Motivation: High acceleration factors place a limit on MRI image reconstruction. This limit is extended to segmentation models when treating these as subsequent independent processes. Goal: Our goal is to produce segmentations directly from sparse k-space measurements without the need for intermediate image reconstruction. Approach: We employ a transformer architecture to encode global k-space information into latent features. The produced latent vectors condition queried coordinates during decoding to generate segmentation class probabilities. Results: The model is able to produce better segmentations across high acceleration factors than image-based segmentation baselines. Impact: Cardiac segmentation directly from undersampled k-space samples circumvents the need for an intermediate image reconstruction step. This allows the potential to assess myocardial structure and function on higher acceleration factors than methods that rely on images as input.
INR meets Multi-Contrast MRI Reconstruction
Sep 05, 2025Abstract:Multi-contrast MRI sequences allow for the acquisition of images with varying tissue contrast within a single scan. The resulting multi-contrast images can be used to extract quantitative information on tissue microstructure. To make such multi-contrast sequences feasible for clinical routine, the usually very long scan times need to be shortened e.g. through undersampling in k-space. However, this comes with challenges for the reconstruction. In general, advanced reconstruction techniques such as compressed sensing or deep learning-based approaches can enable the acquisition of high-quality images despite the acceleration. In this work, we leverage redundant anatomical information of multi-contrast sequences to achieve even higher acceleration rates. We use undersampling patterns that capture the contrast information located at the k-space center, while performing complementary undersampling across contrasts for high frequencies. To reconstruct this highly sparse k-space data, we propose an implicit neural representation (INR) network that is ideal for using the complementary information acquired across contrasts as it jointly reconstructs all contrast images. We demonstrate the benefits of our proposed INR method by applying it to multi-contrast MRI using the MPnRAGE sequence, where it outperforms the state-of-the-art parallel imaging compressed sensing (PICS) reconstruction method, even at higher acceleration factors.
SenseFlow: A Physics-Informed and Self-Ensembling Iterative Framework for Power Flow Estimation
May 18, 2025Abstract:Power flow estimation plays a vital role in ensuring the stability and reliability of electrical power systems, particularly in the context of growing network complexities and renewable energy integration. However, existing studies often fail to adequately address the unique characteristics of power systems, such as the sparsity of network connections and the critical importance of the unique Slack node, which poses significant challenges in achieving high-accuracy estimations. In this paper, we present SenseFlow, a novel physics-informed and self-ensembling iterative framework that integrates two main designs, the Physics-Informed Power Flow Network (FlowNet) and Self-Ensembling Iterative Estimation (SeIter), to carefully address the unique properties of the power system and thereby enhance the power flow estimation. Specifically, SenseFlow enforces the FlowNet to gradually predict high-precision voltage magnitudes and phase angles through the iterative SeIter process. On the one hand, FlowNet employs the Virtual Node Attention and Slack-Gated Feed-Forward modules to facilitate efficient global-local communication in the face of network sparsity and amplify the influence of the Slack node on angle predictions, respectively. On the other hand, SeIter maintains an exponential moving average of FlowNet's parameters to create a robust ensemble model that refines power state predictions throughout the iterative fitting process. Experimental results demonstrate that SenseFlow outperforms existing methods, providing a promising solution for high-accuracy power flow estimation across diverse grid configurations.
Sim-and-Real Co-Training: A Simple Recipe for Vision-Based Robotic Manipulation
Mar 31, 2025Abstract:Large real-world robot datasets hold great potential to train generalist robot models, but scaling real-world human data collection is time-consuming and resource-intensive. Simulation has great potential in supplementing large-scale data, especially with recent advances in generative AI and automated data generation tools that enable scalable creation of robot behavior datasets. However, training a policy solely in simulation and transferring it to the real world often demands substantial human effort to bridge the reality gap. A compelling alternative is to co-train the policy on a mixture of simulation and real-world datasets. Preliminary studies have recently shown this strategy to substantially improve the performance of a policy over one trained on a limited amount of real-world data. Nonetheless, the community lacks a systematic understanding of sim-and-real co-training and what it takes to reap the benefits of simulation data for real-robot learning. This work presents a simple yet effective recipe for utilizing simulation data to solve vision-based robotic manipulation tasks. We derive this recipe from comprehensive experiments that validate the co-training strategy on various simulation and real-world datasets. Using two domains--a robot arm and a humanoid--across diverse tasks, we demonstrate that simulation data can enhance real-world task performance by an average of 38%, even with notable differences between the simulation and real-world data. Videos and additional results can be found at https://co-training.github.io/
Empowering Large Language Models in Wireless Communication: A Novel Dataset and Fine-Tuning Framework
Jan 16, 2025Abstract:In this work, we develop a specialized dataset aimed at enhancing the evaluation and fine-tuning of large language models (LLMs) specifically for wireless communication applications. The dataset includes a diverse set of multi-hop questions, including true/false and multiple-choice types, spanning varying difficulty levels from easy to hard. By utilizing advanced language models for entity extraction and question generation, rigorous data curation processes are employed to maintain high quality and relevance. Additionally, we introduce a Pointwise V-Information (PVI) based fine-tuning method, providing a detailed theoretical analysis and justification for its use in quantifying the information content of training data with 2.24\% and 1.31\% performance boost for different models compared to baselines, respectively. To demonstrate the effectiveness of the fine-tuned models with the proposed methodologies on practical tasks, we also consider different tasks, including summarizing optimization problems from technical papers and solving the mathematical problems related to non-orthogonal multiple access (NOMA), which are generated by using the proposed multi-agent framework. Simulation results show significant performance gain in summarization tasks with 20.9\% in the ROUGE-L metrics. We also study the scaling laws of fine-tuning LLMs and the challenges LLMs face in the field of wireless communications, offering insights into their adaptation to wireless communication tasks. This dataset and fine-tuning methodology aim to enhance the training and evaluation of LLMs, contributing to advancements in LLMs for wireless communication research and applications.
PISCO: Self-Supervised k-Space Regularization for Improved Neural Implicit k-Space Representations of Dynamic MRI
Jan 16, 2025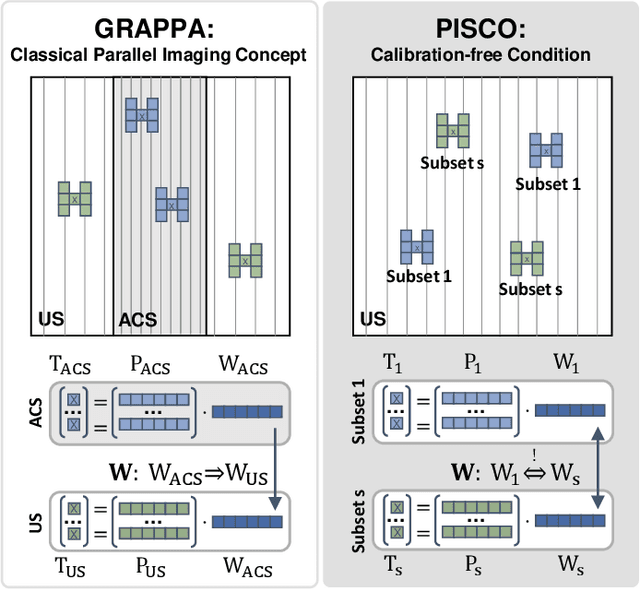
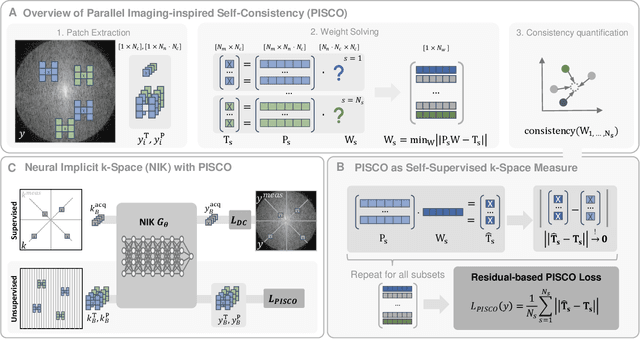
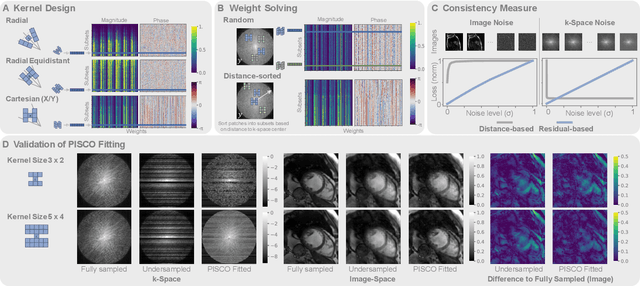
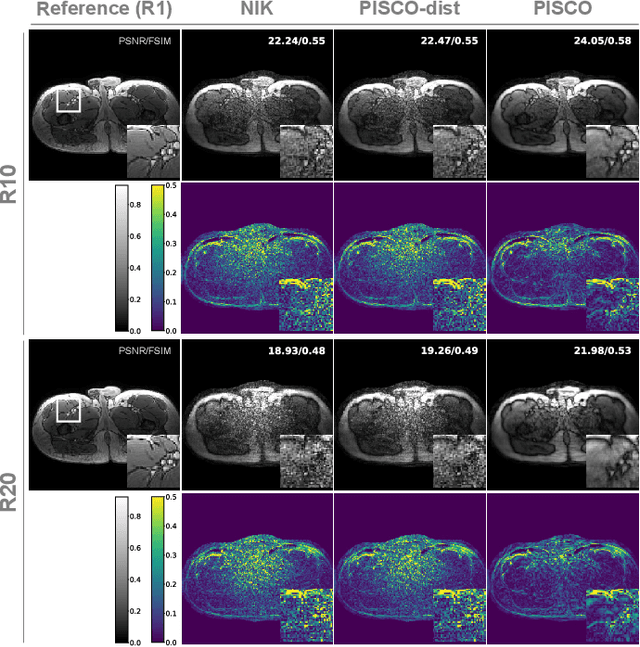
Abstract:Neural implicit k-space representations (NIK) have shown promising results for dynamic magnetic resonance imaging (MRI) at high temporal resolutions. Yet, reducing acquisition time, and thereby available training data, results in severe performance drops due to overfitting. To address this, we introduce a novel self-supervised k-space loss function $\mathcal{L}_\mathrm{PISCO}$, applicable for regularization of NIK-based reconstructions. The proposed loss function is based on the concept of parallel imaging-inspired self-consistency (PISCO), enforcing a consistent global k-space neighborhood relationship without requiring additional data. Quantitative and qualitative evaluations on static and dynamic MR reconstructions show that integrating PISCO significantly improves NIK representations. Particularly for high acceleration factors (R$\geq$54), NIK with PISCO achieves superior spatio-temporal reconstruction quality compared to state-of-the-art methods. Furthermore, an extensive analysis of the loss assumptions and stability shows PISCO's potential as versatile self-supervised k-space loss function for further applications and architectures. Code is available at: https://github.com/compai-lab/2025-pisco-spieker
Subspace Implicit Neural Representations for Real-Time Cardiac Cine MR Imaging
Dec 17, 2024
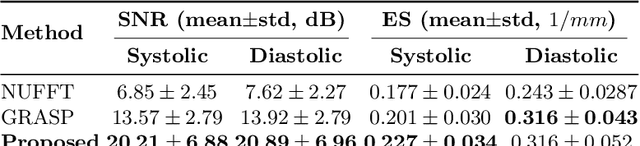
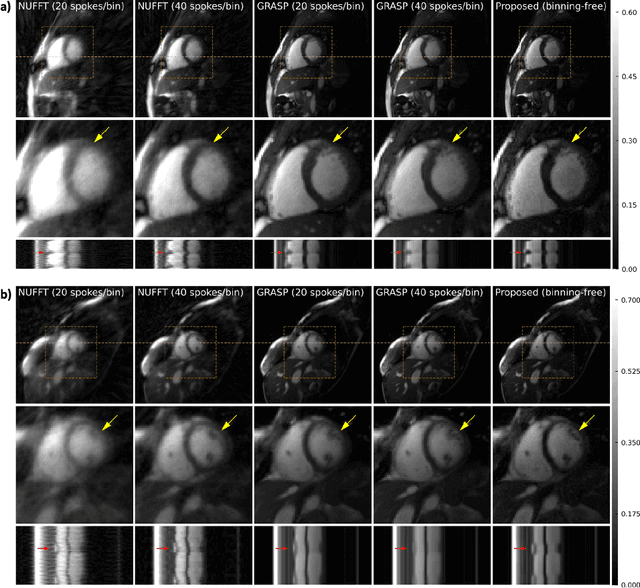
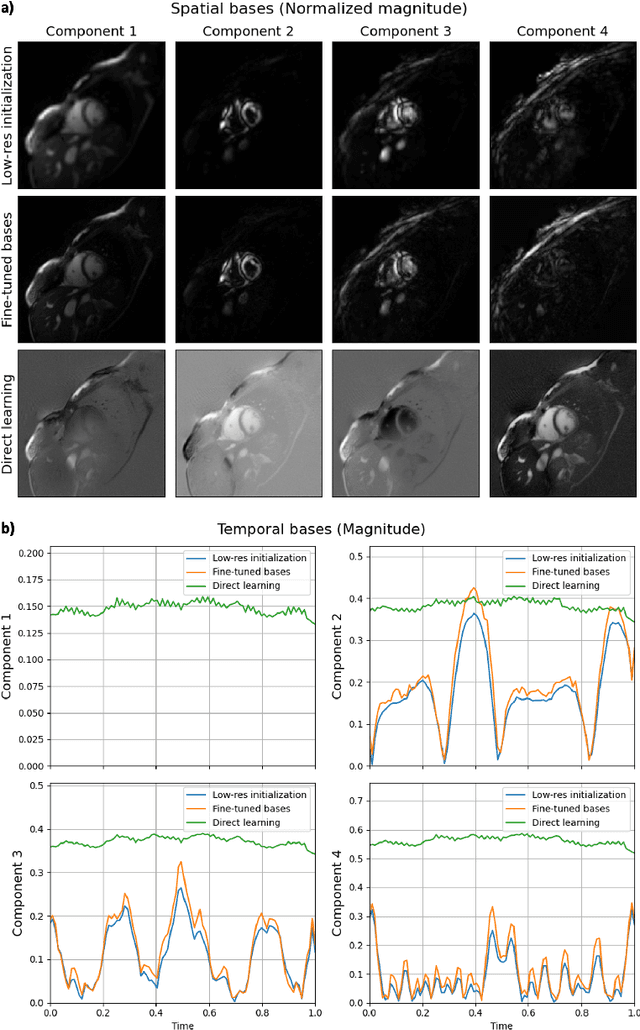
Abstract:Conventional cardiac cine MRI methods rely on retrospective gating, which limits temporal resolution and the ability to capture continuous cardiac dynamics, particularly in patients with arrhythmias and beat-to-beat variations. To address these challenges, we propose a reconstruction framework based on subspace implicit neural representations for real-time cardiac cine MRI of continuously sampled radial data. This approach employs two multilayer perceptrons to learn spatial and temporal subspace bases, leveraging the low-rank properties of cardiac cine MRI. Initialized with low-resolution reconstructions, the networks are fine-tuned using spoke-specific loss functions to recover spatial details and temporal fidelity. Our method directly utilizes the continuously sampled radial k-space spokes during training, thereby eliminating the need for binning and non-uniform FFT. This approach achieves superior spatial and temporal image quality compared to conventional binned methods at the acceleration rate of 10 and 20, demonstrating potential for high-resolution imaging of dynamic cardiac events and enhancing diagnostic capability.
Direct Cardiac Segmentation from Undersampled K-space Using Transformers
May 31, 2024


Abstract:The prevailing deep learning-based methods of predicting cardiac segmentation involve reconstructed magnetic resonance (MR) images. The heavy dependency of segmentation approaches on image quality significantly limits the acceleration rate in fast MR reconstruction. Moreover, the practice of treating reconstruction and segmentation as separate sequential processes leads to artifact generation and information loss in the intermediate stage. These issues pose a great risk to achieving high-quality outcomes. To leverage the redundant k-space information overlooked in this dual-step pipeline, we introduce a novel approach to directly deriving segmentations from sparse k-space samples using a transformer (DiSK). DiSK operates by globally extracting latent features from 2D+time k-space data with attention blocks and subsequently predicting the segmentation label of query points. We evaluate our model under various acceleration factors (ranging from 4 to 64) and compare against two image-based segmentation baselines. Our model consistently outperforms the baselines in Dice and Hausdorff distances across foreground classes for all presented sampling rates.
Self-Supervised k-Space Regularization for Motion-Resolved Abdominal MRI Using Neural Implicit k-Space Representation
Apr 12, 2024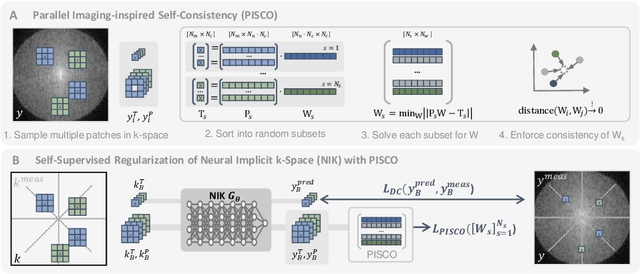
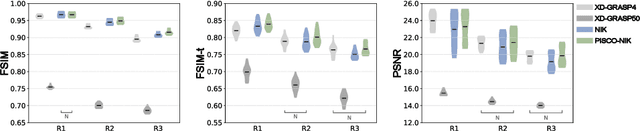
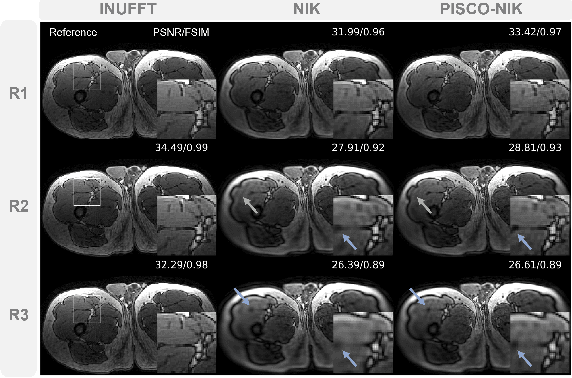
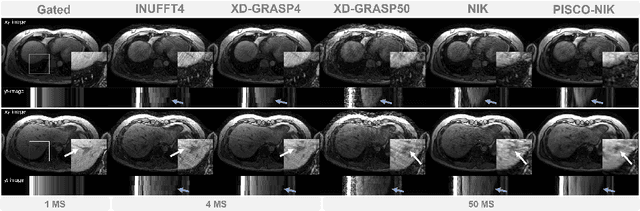
Abstract:Neural implicit k-space representations have shown promising results for dynamic MRI at high temporal resolutions. Yet, their exclusive training in k-space limits the application of common image regularization methods to improve the final reconstruction. In this work, we introduce the concept of parallel imaging-inspired self-consistency (PISCO), which we incorporate as novel self-supervised k-space regularization enforcing a consistent neighborhood relationship. At no additional data cost, the proposed regularization significantly improves neural implicit k-space reconstructions on simulated data. Abdominal in-vivo reconstructions using PISCO result in enhanced spatio-temporal image quality compared to state-of-the-art methods. Code is available at https://github.com/vjspi/PISCO-NIK.
Propagation and Attribution of Uncertainty in Medical Imaging Pipelines
Sep 28, 2023Abstract:Uncertainty estimation, which provides a means of building explainable neural networks for medical imaging applications, have mostly been studied for single deep learning models that focus on a specific task. In this paper, we propose a method to propagate uncertainty through cascades of deep learning models in medical imaging pipelines. This allows us to aggregate the uncertainty in later stages of the pipeline and to obtain a joint uncertainty measure for the predictions of later models. Additionally, we can separately report contributions of the aleatoric, data-based, uncertainty of every component in the pipeline. We demonstrate the utility of our method on a realistic imaging pipeline that reconstructs undersampled brain and knee magnetic resonance (MR) images and subsequently predicts quantitative information from the images, such as the brain volume, or knee side or patient's sex. We quantitatively show that the propagated uncertainty is correlated with input uncertainty and compare the proportions of contributions of pipeline stages to the joint uncertainty measure.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge