Vasiliki Sideri-Lampretsa
CINeMA: Conditional Implicit Neural Multi-Modal Atlas for a Spatio-Temporal Representation of the Perinatal Brain
Jun 11, 2025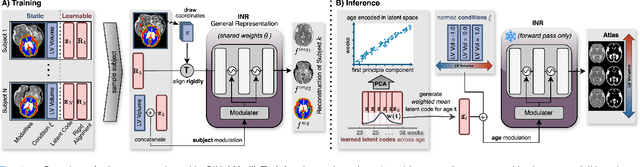
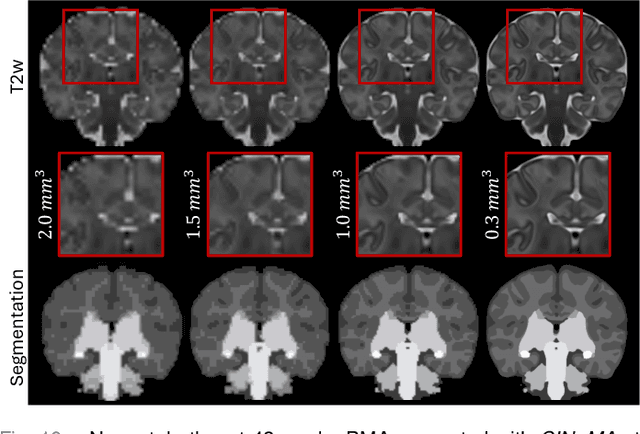
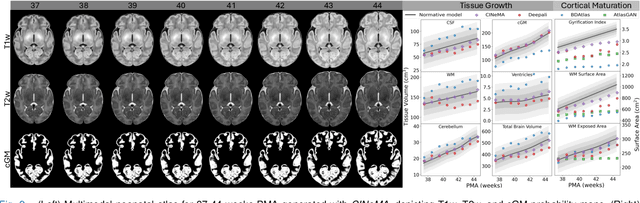
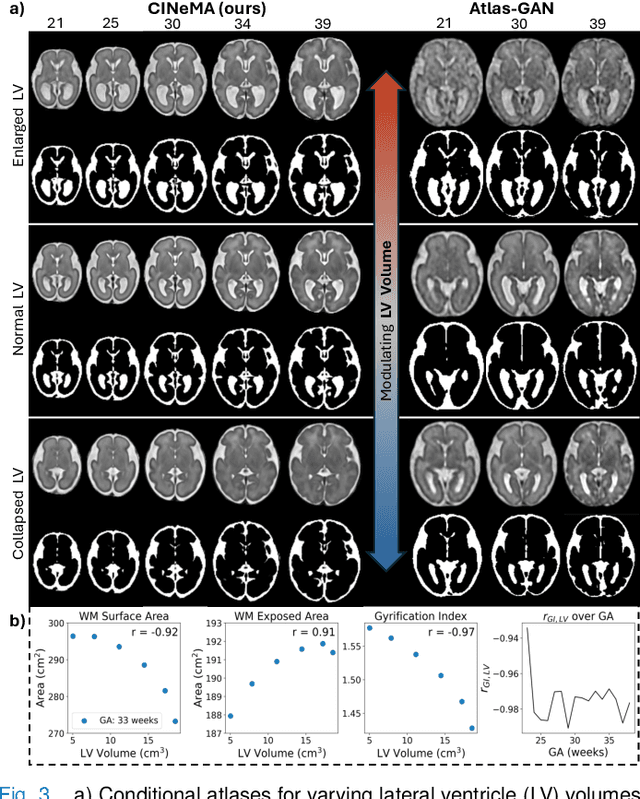
Abstract:Magnetic resonance imaging of fetal and neonatal brains reveals rapid neurodevelopment marked by substantial anatomical changes unfolding within days. Studying this critical stage of the developing human brain, therefore, requires accurate brain models-referred to as atlases-of high spatial and temporal resolution. To meet these demands, established traditional atlases and recently proposed deep learning-based methods rely on large and comprehensive datasets. This poses a major challenge for studying brains in the presence of pathologies for which data remains scarce. We address this limitation with CINeMA (Conditional Implicit Neural Multi-Modal Atlas), a novel framework for creating high-resolution, spatio-temporal, multimodal brain atlases, suitable for low-data settings. Unlike established methods, CINeMA operates in latent space, avoiding compute-intensive image registration and reducing atlas construction times from days to minutes. Furthermore, it enables flexible conditioning on anatomical features including GA, birth age, and pathologies like ventriculomegaly (VM) and agenesis of the corpus callosum (ACC). CINeMA supports downstream tasks such as tissue segmentation and age prediction whereas its generative properties enable synthetic data creation and anatomically informed data augmentation. Surpassing state-of-the-art methods in accuracy, efficiency, and versatility, CINeMA represents a powerful tool for advancing brain research. We release the code and atlases at https://github.com/m-dannecker/CINeMA.
Evaluation of Alignment-Regularity Characteristics in Deformable Image Registration
Mar 10, 2025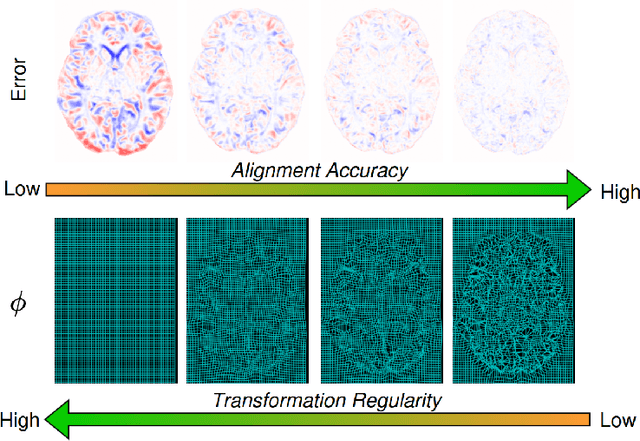
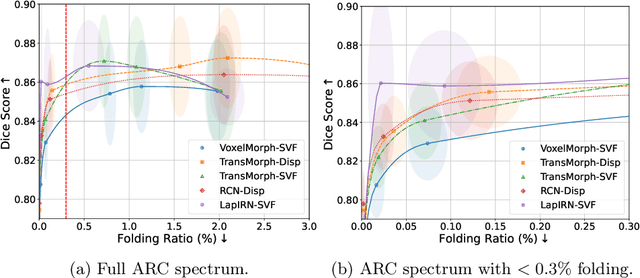
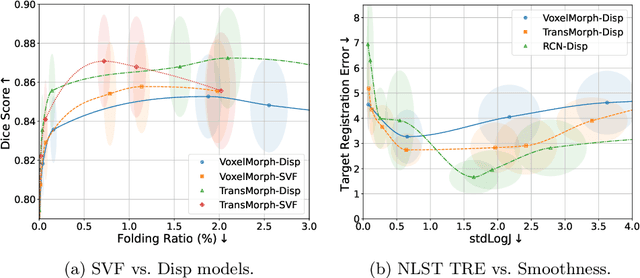
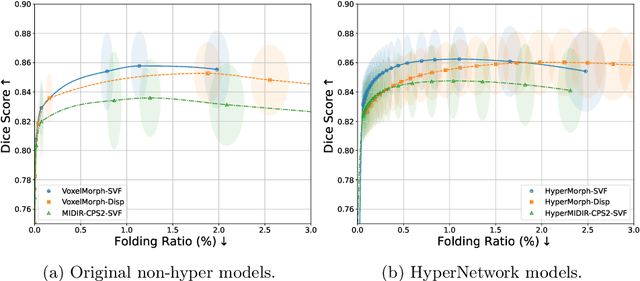
Abstract:Evaluating deformable image registration (DIR) is challenging due to the inherent trade-off between achieving high alignment accuracy and maintaining deformation regularity. In this work, we introduce a novel evaluation scheme based on the alignment-regularity characteristic (ARC) to systematically capture and analyze this trade-off. We first introduce the ARC curves, which describe the performance of a given registration algorithm as a spectrum measured by alignment and regularity metrics. We further adopt a HyperNetwork-based approach that learns to continuously interpolate across the full regularization range, accelerating the construction and improving the sample density of ARC curves. We empirically demonstrate our evaluation scheme using representative learning-based deformable image registration methods with various network architectures and transformation models on two public datasets. We present a range of findings not evident from existing evaluation practices and provide general recommendations for model evaluation and selection using our evaluation scheme. All code relevant is made publicly available.
Deformable Image Registration of Dark-Field Chest Radiographs for Local Lung Signal Change Assessment
Jan 18, 2025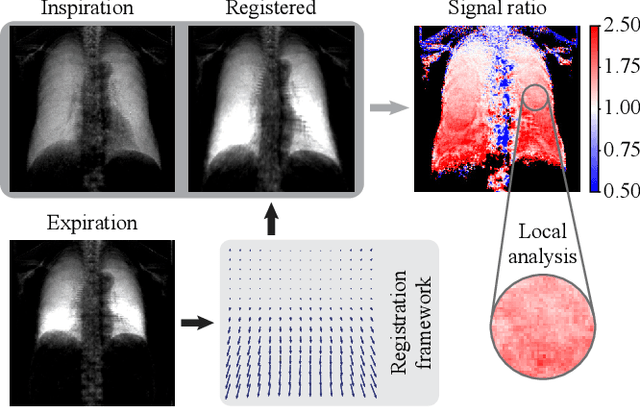
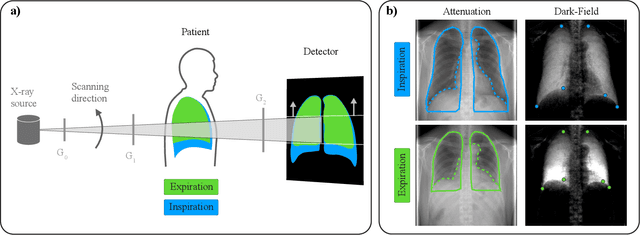
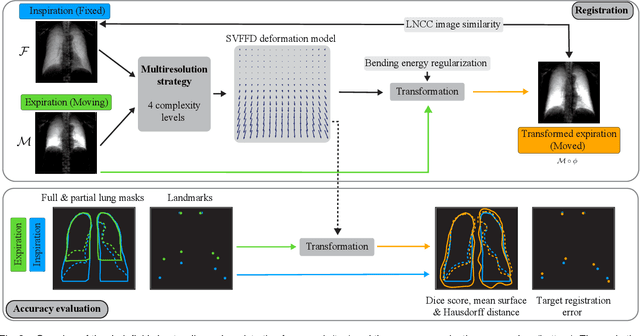
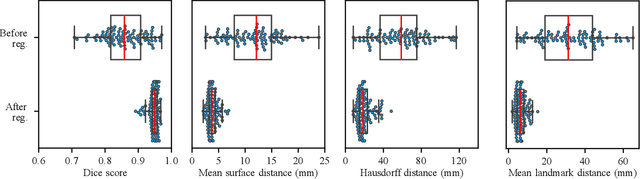
Abstract:Dark-field radiography of the human chest has been demonstrated to have promising potential for the analysis of the lung microstructure and the diagnosis of respiratory diseases. However, previous studies of dark-field chest radiographs evaluated the lung signal only in the inspiratory breathing state. Our work aims to add a new perspective to these previous assessments by locally comparing dark-field lung information between different respiratory states. To this end, we discuss suitable image registration methods for dark-field chest radiographs to enable consistent spatial alignment of the lung in distinct breathing states. Utilizing full inspiration and expiration scans from a clinical chronic obstructive pulmonary disease study, we assess the performance of the proposed registration framework and outline applicable evaluation approaches. Our regional characterization of lung dark-field signal changes between the breathing states provides a proof-of-principle that dynamic radiography-based lung function assessment approaches may benefit from considering registered dark-field images in addition to standard plain chest radiographs.
From Model Based to Learned Regularization in Medical Image Registration: A Comprehensive Review
Dec 20, 2024Abstract:Image registration is fundamental in medical imaging applications, such as disease progression analysis or radiation therapy planning. The primary objective of image registration is to precisely capture the deformation between two or more images, typically achieved by minimizing an optimization problem. Due to its inherent ill-posedness, regularization is a key component in driving the solution toward anatomically meaningful deformations. A wide range of regularization methods has been proposed for both conventional and deep learning-based registration. However, the appropriate application of regularization techniques often depends on the specific registration problem, and no one-fits-all method exists. Despite its importance, regularization is often overlooked or addressed with default approaches, assuming existing methods are sufficient. A comprehensive and structured review remains missing. This review addresses this gap by introducing a novel taxonomy that systematically categorizes the diverse range of proposed regularization methods. It highlights the emerging field of learned regularization, which leverages data-driven techniques to automatically derive deformation properties from the data. Moreover, this review examines the transfer of regularization methods from conventional to learning-based registration, identifies open challenges, and outlines future research directions. By emphasizing the critical role of regularization in image registration, we hope to inspire the research community to reconsider regularization strategies in modern registration algorithms and to explore this rapidly evolving field further.
Image registration is a geometric deep learning task
Dec 17, 2024



Abstract:Data-driven deformable image registration methods predominantly rely on operations that process grid-like inputs. However, applying deformable transformations to an image results in a warped space that deviates from a rigid grid structure. Consequently, data-driven approaches with sequential deformations have to apply grid resampling operations between each deformation step. While artifacts caused by resampling are negligible in high-resolution images, the resampling of sparse, high-dimensional feature grids introduces errors that affect the deformation modeling process. Taking inspiration from Lagrangian reference frames of deformation fields, our work introduces a novel paradigm for data-driven deformable image registration that utilizes geometric deep-learning principles to model deformations without grid requirements. Specifically, we model image features as a set of nodes that freely move in Euclidean space, update their coordinates under graph operations, and dynamically readjust their local neighborhoods. We employ this formulation to construct a multi-resolution deformable registration model, where deformation layers iteratively refine the overall transformation at each resolution without intermediate resampling operations on the feature grids. We investigate our method's ability to fully deformably capture large deformations across a number of medical imaging registration tasks. In particular, we apply our approach (GeoReg) to the registration of inter-subject brain MR images and inhale-exhale lung CT images, showing on par performance with the current state-of-the-art methods. We believe our contribution open up avenues of research to reduce the black-box nature of current learned registration paradigms by explicitly modeling the transformation within the architecture.
General Vision Encoder Features as Guidance in Medical Image Registration
Jul 18, 2024Abstract:General vision encoders like DINOv2 and SAM have recently transformed computer vision. Even though they are trained on natural images, such encoder models have excelled in medical imaging, e.g., in classification, segmentation, and registration. However, no in-depth comparison of different state-of-the-art general vision encoders for medical registration is available. In this work, we investigate how well general vision encoder features can be used in the dissimilarity metrics for medical image registration. We explore two encoders that were trained on natural images as well as one that was fine-tuned on medical data. We apply the features within the well-established B-spline FFD registration framework. In extensive experiments on cardiac cine MRI data, we find that using features as additional guidance for conventional metrics improves the registration quality. The code is available at github.com/compai-lab/2024-miccai-koegl.
Intensity-based 3D motion correction for cardiac MR images
Mar 31, 2024



Abstract:Cardiac magnetic resonance (CMR) image acquisition requires subjects to hold their breath while 2D cine images are acquired. This process assumes that the heart remains in the same position across all slices. However, differences in breathhold positions or patient motion introduce 3D slice misalignments. In this work, we propose an algorithm that simultaneously aligns all SA and LA slices by maximizing the pair-wise intensity agreement between their intersections. Unlike previous works, our approach is formulated as a subject-specific optimization problem and requires no prior knowledge of the underlying anatomy. We quantitatively demonstrate that the proposed method is robust against a large range of rotations and translations by synthetically misaligning 10 motion-free datasets and aligning them back using the proposed method.
Diff-Def: Diffusion-Generated Deformation Fields for Conditional Atlases
Mar 25, 2024



Abstract:Anatomical atlases are widely used for population analysis. Conditional atlases target a particular sub-population defined via certain conditions (e.g. demographics or pathologies) and allow for the investigation of fine-grained anatomical differences - such as morphological changes correlated with age. Existing approaches use either registration-based methods that are unable to handle large anatomical variations or generative models, which can suffer from training instabilities and hallucinations. To overcome these limitations, we use latent diffusion models to generate deformation fields, which transform a general population atlas into one representing a specific sub-population. By generating a deformation field and registering the conditional atlas to a neighbourhood of images, we ensure structural plausibility and avoid hallucinations, which can occur during direct image synthesis. We compare our method to several state-of-the-art atlas generation methods in experiments using 5000 brain as well as whole-body MR images from UK Biobank. Our method generates highly realistic atlases with smooth transformations and high anatomical fidelity, outperforming the baselines.
MAD: Modality Agnostic Distance Measure for Image Registration
Sep 06, 2023Abstract:Multi-modal image registration is a crucial pre-processing step in many medical applications. However, it is a challenging task due to the complex intensity relationships between different imaging modalities, which can result in large discrepancy in image appearance. The success of multi-modal image registration, whether it is conventional or learning based, is predicated upon the choice of an appropriate distance (or similarity) measure. Particularly, deep learning registration algorithms lack in accuracy or even fail completely when attempting to register data from an "unseen" modality. In this work, we present Modality Agnostic Distance (MAD), a deep image distance}] measure that utilises random convolutions to learn the inherent geometry of the images while being robust to large appearance changes. Random convolutions are geometry-preserving modules which we use to simulate an infinite number of synthetic modalities alleviating the need for aligned paired data during training. We can therefore train MAD on a mono-modal dataset and successfully apply it to a multi-modal dataset. We demonstrate that not only can MAD affinely register multi-modal images successfully, but it has also a larger capture range than traditional measures such as Mutual Information and Normalised Gradient Fields.
Constructing Population-Specific Atlases from Whole Body MRI: Application to the UKBB
Aug 28, 2023Abstract:Population atlases are commonly utilised in medical imaging to facilitate the investigation of variability across populations. Such atlases enable the mapping of medical images into a common coordinate system, promoting comparability and enabling the study of inter-subject differences. Constructing such atlases becomes particularly challenging when working with highly heterogeneous datasets, such as whole-body images, where subjects show significant anatomical variations. In this work, we propose a pipeline for generating a standardised whole-body atlas for a highly heterogeneous population by partitioning the population into meaningful subgroups. We create six whole-body atlases that represent a healthy population average using magnetic resonance (MR) images from the UK Biobank dataset. We furthermore unbias them, and this way obtain a realistic representation of the population. In addition to the anatomical atlases, we generate probabilistic atlases that capture the distributions of abdominal fat and five abdominal organs across the population. We demonstrate different applications of these atlases, using the differences between subjects with medical conditions such as diabetes and cardiovascular diseases and healthy subjects from the atlas space. With this work, we make the constructed anatomical and label atlases publically available and anticipate them to support medical research conducted on whole-body MR images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge