Mengyun Qiao
Multimodal Conditional MeshGAN for Personalized Aneurysm Growth Prediction
Aug 27, 2025Abstract:Personalized, accurate prediction of aortic aneurysm progression is essential for timely intervention but remains challenging due to the need to model both subtle local deformations and global anatomical changes within complex 3D geometries. We propose MCMeshGAN, the first multimodal conditional mesh-to-mesh generative adversarial network for 3D aneurysm growth prediction. MCMeshGAN introduces a dual-branch architecture combining a novel local KNN-based convolutional network (KCN) to preserve fine-grained geometric details and a global graph convolutional network (GCN) to capture long-range structural context, overcoming the over-smoothing limitations of deep GCNs. A dedicated condition branch encodes clinical attributes (age, sex) and the target time interval to generate anatomically plausible, temporally controlled predictions, enabling retrospective and prospective modeling. We curated TAAMesh, a new longitudinal thoracic aortic aneurysm mesh dataset consisting of 590 multimodal records (CT scans, 3D meshes, and clinical data) from 208 patients. Extensive experiments demonstrate that MCMeshGAN consistently outperforms state-of-the-art baselines in both geometric accuracy and clinically important diameter estimation. This framework offers a robust step toward clinically deployable, personalized 3D disease trajectory modeling. The source code for MCMeshGAN and the baseline methods is publicly available at https://github.com/ImperialCollegeLondon/MCMeshGAN.
A Personalised 3D+t Mesh Generative Model for Unveiling Normal Heart Dynamics
Sep 20, 2024



Abstract:Understanding the structure and motion of the heart is crucial for diagnosing and managing cardiovascular diseases, the leading cause of global death. There is wide variation in cardiac shape and motion patterns, that are influenced by demographic, anthropometric and disease factors. Unravelling the normal patterns of shape and motion, as well as understanding how each individual deviates from the norm, would facilitate accurate diagnosis and personalised treatment strategies. To this end, we developed a novel conditional generative model, MeshHeart, to learn the distribution of cardiac shape and motion patterns. MeshHeart is capable of generating 3D+t cardiac mesh sequences, taking into account clinical factors such as age, sex, weight and height. To model the high-dimensional and complex spatio-temporal mesh data, MeshHeart employs a geometric encoder to represent cardiac meshes in a latent space, followed by a temporal Transformer to model the motion dynamics of latent representations. Based on MeshHeart, we investigate the latent space of 3D+t cardiac mesh sequences and propose a novel distance metric termed latent delta, which quantifies the deviation of a real heart from its personalised normative pattern in the latent space. In experiments using a large dataset of 38,309 subjects, MeshHeart demonstrates a high performance in cardiac mesh sequence reconstruction and generation. Features defined in the latent space are highly discriminative for cardiac disease classification, whereas the latent delta exhibits strong correlation with clinical phenotypes in phenome-wide association studies. The codes and models of this study will be released to benefit further research on digital heart modelling.
A Foundation Model for Brain Lesion Segmentation with Mixture of Modality Experts
May 16, 2024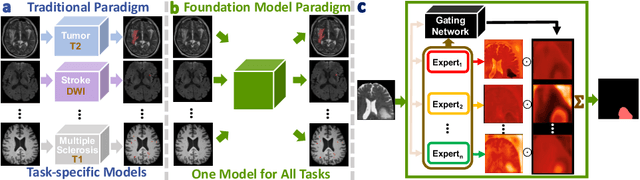
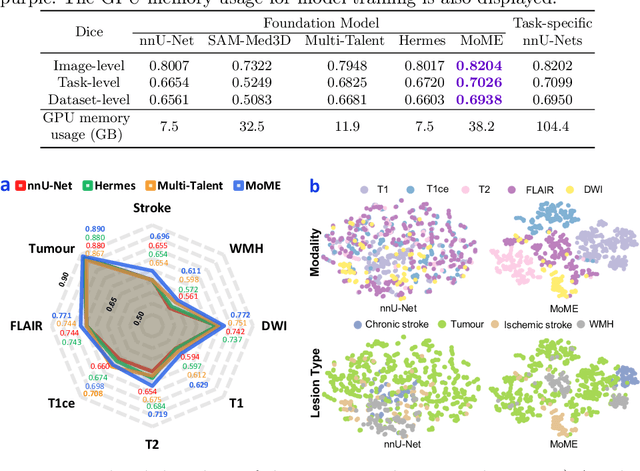

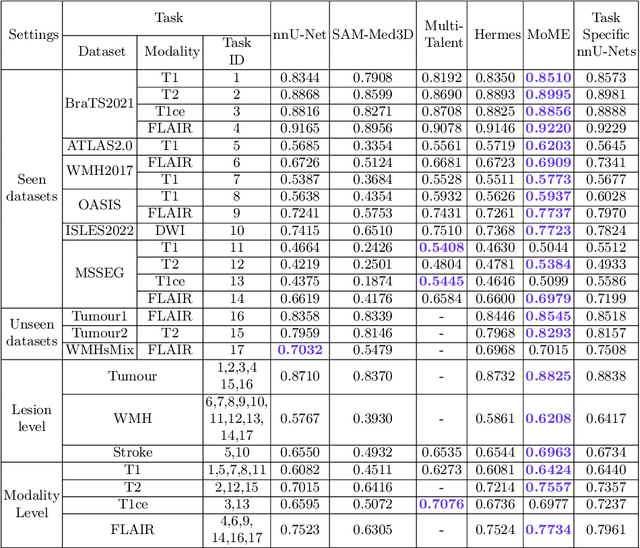
Abstract:Brain lesion segmentation plays an essential role in neurological research and diagnosis. As brain lesions can be caused by various pathological alterations, different types of brain lesions tend to manifest with different characteristics on different imaging modalities. Due to this complexity, brain lesion segmentation methods are often developed in a task-specific manner. A specific segmentation model is developed for a particular lesion type and imaging modality. However, the use of task-specific models requires predetermination of the lesion type and imaging modality, which complicates their deployment in real-world scenarios. In this work, we propose a universal foundation model for 3D brain lesion segmentation, which can automatically segment different types of brain lesions for input data of various imaging modalities. We formulate a novel Mixture of Modality Experts (MoME) framework with multiple expert networks attending to different imaging modalities. A hierarchical gating network combines the expert predictions and fosters expertise collaboration. Furthermore, we introduce a curriculum learning strategy during training to avoid the degeneration of each expert network and preserve their specialization. We evaluated the proposed method on nine brain lesion datasets, encompassing five imaging modalities and eight lesion types. The results show that our model outperforms state-of-the-art universal models and provides promising generalization to unseen datasets.
T1/T2 relaxation temporal modelling from accelerated acquisitions using a Latent Transformer
Sep 28, 2023Abstract:Quantitative cardiac magnetic resonance T1 and T2 mapping enable myocardial tissue characterisation but the lengthy scan times restrict their widespread clinical application. We propose a deep learning method that incorporates a time dependency Latent Transformer module to model relationships between parameterised time frames for improved reconstruction from undersampled data. The module, implemented as a multi-resolution sequence-to-sequence transformer, is integrated into an encoder-decoder architecture to leverage the inherent temporal correlations in relaxation processes. The presented results for accelerated T1 and T2 mapping show the model recovers maps with higher fidelity by explicit incorporation of time dynamics. This work demonstrates the importance of temporal modelling for artifact-free reconstruction in quantitative MRI.
LesionMix: A Lesion-Level Data Augmentation Method for Medical Image Segmentation
Aug 17, 2023



Abstract:Data augmentation has become a de facto component of deep learning-based medical image segmentation methods. Most data augmentation techniques used in medical imaging focus on spatial and intensity transformations to improve the diversity of training images. They are often designed at the image level, augmenting the full image, and do not pay attention to specific abnormalities within the image. Here, we present LesionMix, a novel and simple lesion-aware data augmentation method. It performs augmentation at the lesion level, increasing the diversity of lesion shape, location, intensity and load distribution, and allowing both lesion populating and inpainting. Experiments on different modalities and different lesion datasets, including four brain MR lesion datasets and one liver CT lesion dataset, demonstrate that LesionMix achieves promising performance in lesion image segmentation, outperforming several recent Mix-based data augmentation methods. The code will be released at https://github.com/dogabasaran/lesionmix.
M-FLAG: Medical Vision-Language Pre-training with Frozen Language Models and Latent Space Geometry Optimization
Jul 19, 2023
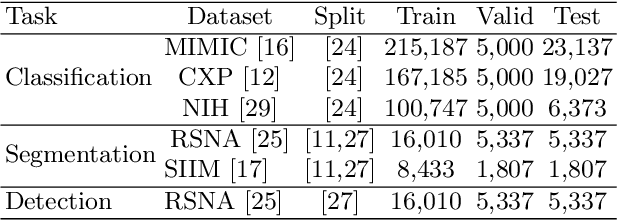
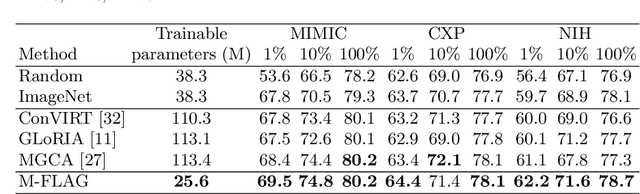

Abstract:Medical vision-language models enable co-learning and integrating features from medical imaging and clinical text. However, these models are not easy to train and the latent representation space can be complex. Here we propose a novel way for pre-training and regularising medical vision-language models. The proposed method, named Medical vision-language pre-training with Frozen language models and Latent spAce Geometry optimization (M-FLAG), leverages a frozen language model for training stability and efficiency and introduces a novel orthogonality loss to harmonize the latent space geometry. We demonstrate the potential of the pre-trained model on three downstream tasks: medical image classification, segmentation, and object detection. Extensive experiments across five public datasets demonstrate that M-FLAG significantly outperforms existing medical vision-language pre-training approaches and reduces the number of parameters by 78\%. Notably, M-FLAG achieves outstanding performance on the segmentation task while using only 1\% of the RSNA dataset, even outperforming ImageNet pre-trained models that have been fine-tuned using 100\% of the data.
Feature-Conditioned Cascaded Video Diffusion Models for Precise Echocardiogram Synthesis
Mar 23, 2023Abstract:Image synthesis is expected to provide value for the translation of machine learning methods into clinical practice. Fundamental problems like model robustness, domain transfer, causal modelling, and operator training become approachable through synthetic data. Especially, heavily operator-dependant modalities like Ultrasound imaging require robust frameworks for image and video generation. So far, video generation has only been possible by providing input data that is as rich as the output data, e.g., image sequence plus conditioning in, video out. However, clinical documentation is usually scarce and only single images are reported and stored, thus retrospective patient-specific analysis or the generation of rich training data becomes impossible with current approaches. In this paper, we extend elucidated diffusion models for video modelling to generate plausible video sequences from single images and arbitrary conditioning with clinical parameters. We explore this idea within the context of echocardiograms by looking into the variation of the Left Ventricle Ejection Fraction, the most essential clinical metric gained from these examinations. We use the publicly available EchoNet-Dynamic dataset for all our experiments. Our image to sequence approach achieves an $R^2$ score of 93%, which is 38 points higher than recently proposed sequence to sequence generation methods. Code and models will be available at: https://github.com/HReynaud/EchoDiffusion.
CHeart: A Conditional Spatio-Temporal Generative Model for Cardiac Anatomy
Jan 30, 2023Abstract:Two key questions in cardiac image analysis are to assess the anatomy and motion of the heart from images; and to understand how they are associated with non-imaging clinical factors such as gender, age and diseases. While the first question can often be addressed by image segmentation and motion tracking algorithms, our capability to model and to answer the second question is still limited. In this work, we propose a novel conditional generative model to describe the 4D spatio-temporal anatomy of the heart and its interaction with non-imaging clinical factors. The clinical factors are integrated as the conditions of the generative modelling, which allows us to investigate how these factors influence the cardiac anatomy. We evaluate the model performance in mainly two tasks, anatomical sequence completion and sequence generation. The model achieves a high performance in anatomical sequence completion, comparable to or outperforming other state-of-the-art generative models. In terms of sequence generation, given clinical conditions, the model can generate realistic synthetic 4D sequential anatomies that share similar distributions with the real data.
Generative Modelling of the Ageing Heart with Cross-Sectional Imaging and Clinical Data
Aug 28, 2022



Abstract:Cardiovascular disease, the leading cause of death globally, is an age-related disease. Understanding the morphological and functional changes of the heart during ageing is a key scientific question, the answer to which will help us define important risk factors of cardiovascular disease and monitor disease progression. In this work, we propose a novel conditional generative model to describe the changes of 3D anatomy of the heart during ageing. The proposed model is flexible and allows integration of multiple clinical factors (e.g. age, gender) into the generating process. We train the model on a large-scale cross-sectional dataset of cardiac anatomies and evaluate on both cross-sectional and longitudinal datasets. The model demonstrates excellent performance in predicting the longitudinal evolution of the ageing heart and modelling its data distribution.
Subject-Specific Lesion Generation and Pseudo-Healthy Synthesis for Multiple Sclerosis Brain Images
Aug 03, 2022



Abstract:Understanding the intensity characteristics of brain lesions is key for defining image-based biomarkers in neurological studies and for predicting disease burden and outcome. In this work, we present a novel foreground-based generative method for modelling the local lesion characteristics that can both generate synthetic lesions on healthy images and synthesize subject-specific pseudo-healthy images from pathological images. Furthermore, the proposed method can be used as a data augmentation module to generate synthetic images for training brain image segmentation networks. Experiments on multiple sclerosis (MS) brain images acquired on magnetic resonance imaging (MRI) demonstrate that the proposed method can generate highly realistic pseudo-healthy and pseudo-pathological brain images. Data augmentation using the synthetic images improves the brain image segmentation performance compared to traditional data augmentation methods as well as a recent lesion-aware data augmentation technique, CarveMix. The code will be released at https://github.com/dogabasaran/lesion-synthesis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge