Mischa Dombrowski
LCMem: A Universal Model for Robust Image Memorization Detection
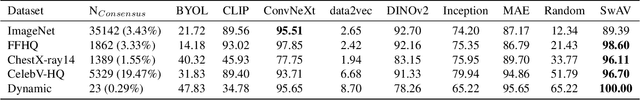
Dec 16, 2025Abstract:Recent advances in generative image modeling have achieved visual realism sufficient to deceive human experts, yet their potential for privacy preserving data sharing remains insufficiently understood. A central obstacle is the absence of reliable memorization detection mechanisms, limited quantitative evaluation, and poor generalization of existing privacy auditing methods across domains. To address this, we propose to view memorization detection as a unified problem at the intersection of re-identification and copy detection, whose complementary goals cover both identity consistency and augmentation-robust duplication, and introduce Latent Contrastive Memorization Network (LCMem), a cross-domain model evaluated jointly on both tasks. LCMem achieves this through a two-stage training strategy that first learns identity consistency before incorporating augmentation-robust copy detection. Across six benchmark datasets, LCMem achieves improvements of up to 16 percentage points on re-identification and 30 percentage points on copy detection, enabling substantially more reliable memorization detection at scale. Our results show that existing privacy filters provide limited performance and robustness, highlighting the need for stronger protection mechanisms. We show that LCMem sets a new standard for cross-domain privacy auditing, offering reliable and scalable memorization detection. Code and model is publicly available at https://github.com/MischaD/LCMem.
Graph Conditioned Diffusion for Controllable Histopathology Image Generation
Oct 08, 2025Abstract:Recent advances in Diffusion Probabilistic Models (DPMs) have set new standards in high-quality image synthesis. Yet, controlled generation remains challenging, particularly in sensitive areas such as medical imaging. Medical images feature inherent structure such as consistent spatial arrangement, shape or texture, all of which are critical for diagnosis. However, existing DPMs operate in noisy latent spaces that lack semantic structure and strong priors, making it difficult to ensure meaningful control over generated content. To address this, we propose graph-based object-level representations for Graph-Conditioned-Diffusion. Our approach generates graph nodes corresponding to each major structure in the image, encapsulating their individual features and relationships. These graph representations are processed by a transformer module and integrated into a diffusion model via the text-conditioning mechanism, enabling fine-grained control over generation. We evaluate this approach using a real-world histopathology use case, demonstrating that our generated data can reliably substitute for annotated patient data in downstream segmentation tasks. The code is available here.
Ontology-Based Concept Distillation for Radiology Report Retrieval and Labeling
Aug 27, 2025Abstract:Retrieval-augmented learning based on radiology reports has emerged as a promising direction to improve performance on long-tail medical imaging tasks, such as rare disease detection in chest X-rays. Most existing methods rely on comparing high-dimensional text embeddings from models like CLIP or CXR-BERT, which are often difficult to interpret, computationally expensive, and not well-aligned with the structured nature of medical knowledge. We propose a novel, ontology-driven alternative for comparing radiology report texts based on clinically grounded concepts from the Unified Medical Language System (UMLS). Our method extracts standardised medical entities from free-text reports using an enhanced pipeline built on RadGraph-XL and SapBERT. These entities are linked to UMLS concepts (CUIs), enabling a transparent, interpretable set-based representation of each report. We then define a task-adaptive similarity measure based on a modified and weighted version of the Tversky Index that accounts for synonymy, negation, and hierarchical relationships between medical entities. This allows efficient and semantically meaningful similarity comparisons between reports. We demonstrate that our approach outperforms state-of-the-art embedding-based retrieval methods in a radiograph classification task on MIMIC-CXR, particularly in long-tail settings. Additionally, we use our pipeline to generate ontology-backed disease labels for MIMIC-CXR, offering a valuable new resource for downstream learning tasks. Our work provides more explainable, reliable, and task-specific retrieval strategies in clinical AI systems, especially when interpretability and domain knowledge integration are essential. Our code is available at https://github.com/Felix-012/ontology-concept-distillation
Generate to Ground: Multimodal Text Conditioning Boosts Phrase Grounding in Medical Vision-Language Models
Jul 16, 2025Abstract:Phrase grounding, i.e., mapping natural language phrases to specific image regions, holds significant potential for disease localization in medical imaging through clinical reports. While current state-of-the-art methods rely on discriminative, self-supervised contrastive models, we demonstrate that generative text-to-image diffusion models, leveraging cross-attention maps, can achieve superior zero-shot phrase grounding performance. Contrary to prior assumptions, we show that fine-tuning diffusion models with a frozen, domain-specific language model, such as CXR-BERT, substantially outperforms domain-agnostic counterparts. This setup achieves remarkable improvements, with mIoU scores doubling those of current discriminative methods. These findings highlight the underexplored potential of generative models for phrase grounding tasks. To further enhance performance, we introduce Bimodal Bias Merging (BBM), a novel post-processing technique that aligns text and image biases to identify regions of high certainty. BBM refines cross-attention maps, achieving even greater localization accuracy. Our results establish generative approaches as a more effective paradigm for phrase grounding in the medical imaging domain, paving the way for more robust and interpretable applications in clinical practice. The source code and model weights are available at https://github.com/Felix-012/generate_to_ground.
Video Dataset Condensation with Diffusion Models
May 10, 2025Abstract:In recent years, the rapid expansion of dataset sizes and the increasing complexity of deep learning models have significantly escalated the demand for computational resources, both for data storage and model training. Dataset distillation has emerged as a promising solution to address this challenge by generating a compact synthetic dataset that retains the essential information from a large real dataset. However, existing methods often suffer from limited performance and poor data quality, particularly in the video domain. In this paper, we focus on video dataset distillation by employing a video diffusion model to generate high-quality synthetic videos. To enhance representativeness, we introduce Video Spatio-Temporal U-Net (VST-UNet), a model designed to select a diverse and informative subset of videos that effectively captures the characteristics of the original dataset. To further optimize computational efficiency, we explore a training-free clustering algorithm, Temporal-Aware Cluster-based Distillation (TAC-DT), to select representative videos without requiring additional training overhead. We validate the effectiveness of our approach through extensive experiments on four benchmark datasets, demonstrating performance improvements of up to \(10.61\%\) over the state-of-the-art. Our method consistently outperforms existing approaches across all datasets, establishing a new benchmark for video dataset distillation.
Image Generation Diversity Issues and How to Tame Them
Nov 25, 2024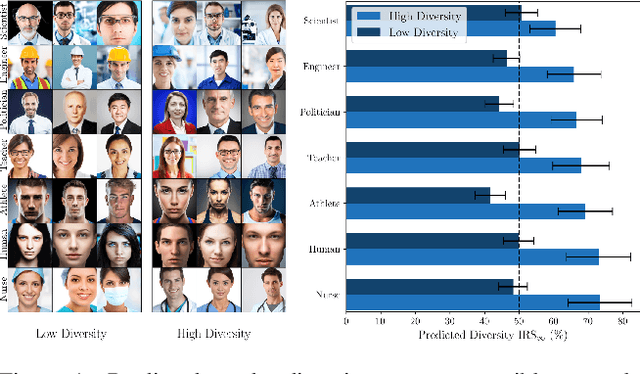
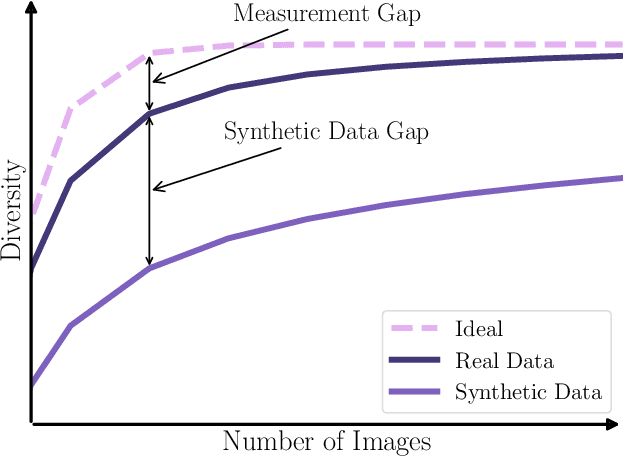
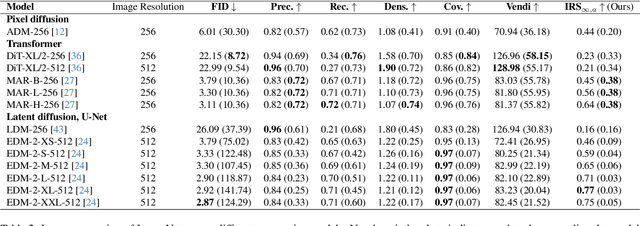
Abstract:Generative methods now produce outputs nearly indistinguishable from real data but often fail to fully capture the data distribution. Unlike quality issues, diversity limitations in generative models are hard to detect visually, requiring specific metrics for assessment. In this paper, we draw attention to the current lack of diversity in generative models and the inability of common metrics to measure this. We achieve this by framing diversity as an image retrieval problem, where we measure how many real images can be retrieved using synthetic data as queries. This yields the Image Retrieval Score (IRS), an interpretable, hyperparameter-free metric that quantifies the diversity of a generative model's output. IRS requires only a subset of synthetic samples and provides a statistical measure of confidence. Our experiments indicate that current feature extractors commonly used in generative model assessment are inadequate for evaluating diversity effectively. Consequently, we perform an extensive search for the best feature extractors to assess diversity. Evaluation reveals that current diffusion models converge to limited subsets of the real distribution, with no current state-of-the-art models superpassing 77% of the diversity of the training data. To address this limitation, we introduce Diversity-Aware Diffusion Models (DiADM), a novel approach that improves diversity of unconditional diffusion models without loss of image quality. We do this by disentangling diversity from image quality by using a diversity aware module that uses pseudo-unconditional features as input. We provide a Python package offering unified feature extraction and metric computation to further facilitate the evaluation of generative models https://github.com/MischaD/beyondfid.
Uncovering Hidden Subspaces in Video Diffusion Models Using Re-Identification
Nov 07, 2024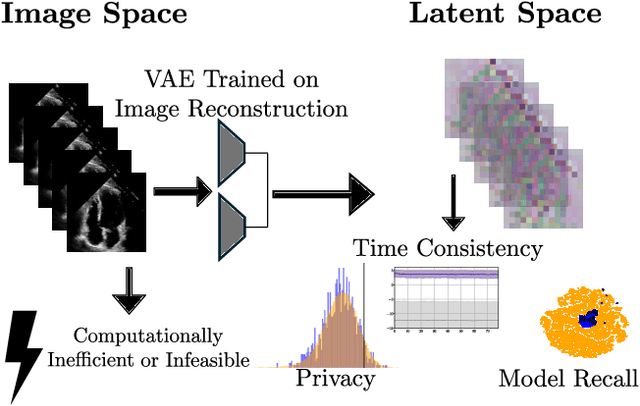
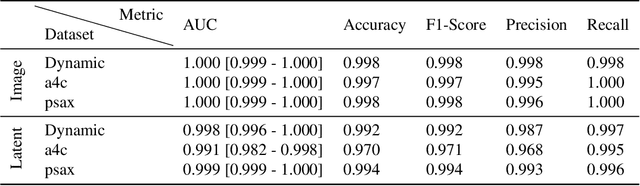
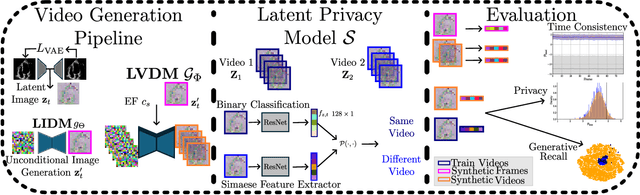
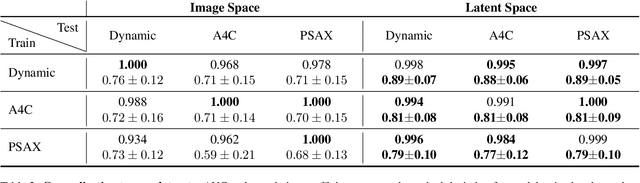
Abstract:Latent Video Diffusion Models can easily deceive casual observers and domain experts alike thanks to the produced image quality and temporal consistency. Beyond entertainment, this creates opportunities around safe data sharing of fully synthetic datasets, which are crucial in healthcare, as well as other domains relying on sensitive personal information. However, privacy concerns with this approach have not fully been addressed yet, and models trained on synthetic data for specific downstream tasks still perform worse than those trained on real data. This discrepancy may be partly due to the sampling space being a subspace of the training videos, effectively reducing the training data size for downstream models. Additionally, the reduced temporal consistency when generating long videos could be a contributing factor. In this paper, we first show that training privacy-preserving models in latent space is computationally more efficient and generalize better. Furthermore, to investigate downstream degradation factors, we propose to use a re-identification model, previously employed as a privacy preservation filter. We demonstrate that it is sufficient to train this model on the latent space of the video generator. Subsequently, we use these models to evaluate the subspace covered by synthetic video datasets and thus introduce a new way to measure the faithfulness of generative machine learning models. We focus on a specific application in healthcare echocardiography to illustrate the effectiveness of our novel methods. Our findings indicate that only up to 30.8% of the training videos are learned in latent video diffusion models, which could explain the lack of performance when training downstream tasks on synthetic data.
JVID: Joint Video-Image Diffusion for Visual-Quality and Temporal-Consistency in Video Generation
Sep 21, 2024



Abstract:We introduce the Joint Video-Image Diffusion model (JVID), a novel approach to generating high-quality and temporally coherent videos. We achieve this by integrating two diffusion models: a Latent Image Diffusion Model (LIDM) trained on images and a Latent Video Diffusion Model (LVDM) trained on video data. Our method combines these models in the reverse diffusion process, where the LIDM enhances image quality and the LVDM ensures temporal consistency. This unique combination allows us to effectively handle the complex spatio-temporal dynamics in video generation. Our results demonstrate quantitative and qualitative improvements in producing realistic and coherent videos.
EchoNet-Synthetic: Privacy-preserving Video Generation for Safe Medical Data Sharing
Jun 02, 2024Abstract:To make medical datasets accessible without sharing sensitive patient information, we introduce a novel end-to-end approach for generative de-identification of dynamic medical imaging data. Until now, generative methods have faced constraints in terms of fidelity, spatio-temporal coherence, and the length of generation, failing to capture the complete details of dataset distributions. We present a model designed to produce high-fidelity, long and complete data samples with near-real-time efficiency and explore our approach on a challenging task: generating echocardiogram videos. We develop our generation method based on diffusion models and introduce a protocol for medical video dataset anonymization. As an exemplar, we present EchoNet-Synthetic, a fully synthetic, privacy-compliant echocardiogram dataset with paired ejection fraction labels. As part of our de-identification protocol, we evaluate the quality of the generated dataset and propose to use clinical downstream tasks as a measurement on top of widely used but potentially biased image quality metrics. Experimental outcomes demonstrate that EchoNet-Synthetic achieves comparable dataset fidelity to the actual dataset, effectively supporting the ejection fraction regression task. Code, weights and dataset are available at https://github.com/HReynaud/EchoNet-Synthetic.
Many tasks make light work: Learning to localise medical anomalies from multiple synthetic tasks
Jul 03, 2023Abstract:There is a growing interest in single-class modelling and out-of-distribution detection as fully supervised machine learning models cannot reliably identify classes not included in their training. The long tail of infinitely many out-of-distribution classes in real-world scenarios, e.g., for screening, triage, and quality control, means that it is often necessary to train single-class models that represent an expected feature distribution, e.g., from only strictly healthy volunteer data. Conventional supervised machine learning would require the collection of datasets that contain enough samples of all possible diseases in every imaging modality, which is not realistic. Self-supervised learning methods with synthetic anomalies are currently amongst the most promising approaches, alongside generative auto-encoders that analyse the residual reconstruction error. However, all methods suffer from a lack of structured validation, which makes calibration for deployment difficult and dataset-dependant. Our method alleviates this by making use of multiple visually-distinct synthetic anomaly learning tasks for both training and validation. This enables more robust training and generalisation. With our approach we can readily outperform state-of-the-art methods, which we demonstrate on exemplars in brain MRI and chest X-rays. Code is available at https://github.com/matt-baugh/many-tasks-make-light-work .
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge