Sarah Cechnicka
Graph Conditioned Diffusion for Controllable Histopathology Image Generation
Oct 08, 2025Abstract:Recent advances in Diffusion Probabilistic Models (DPMs) have set new standards in high-quality image synthesis. Yet, controlled generation remains challenging, particularly in sensitive areas such as medical imaging. Medical images feature inherent structure such as consistent spatial arrangement, shape or texture, all of which are critical for diagnosis. However, existing DPMs operate in noisy latent spaces that lack semantic structure and strong priors, making it difficult to ensure meaningful control over generated content. To address this, we propose graph-based object-level representations for Graph-Conditioned-Diffusion. Our approach generates graph nodes corresponding to each major structure in the image, encapsulating their individual features and relationships. These graph representations are processed by a transformer module and integrated into a diffusion model via the text-conditioning mechanism, enabling fine-grained control over generation. We evaluate this approach using a real-world histopathology use case, demonstrating that our generated data can reliably substitute for annotated patient data in downstream segmentation tasks. The code is available here.
Dataset Distillation with Probabilistic Latent Features
May 10, 2025Abstract:As deep learning models grow in complexity and the volume of training data increases, reducing storage and computational costs becomes increasingly important. Dataset distillation addresses this challenge by synthesizing a compact set of synthetic data that can effectively replace the original dataset in downstream classification tasks. While existing methods typically rely on mapping data from pixel space to the latent space of a generative model, we propose a novel stochastic approach that models the joint distribution of latent features. This allows our method to better capture spatial structures and produce diverse synthetic samples, which benefits model training. Specifically, we introduce a low-rank multivariate normal distribution parameterized by a lightweight network. This design maintains low computational complexity and is compatible with various matching networks used in dataset distillation. After distillation, synthetic images are generated by feeding the learned latent features into a pretrained generator. These synthetic images are then used to train classification models, and performance is evaluated on real test set. We validate our method on several benchmarks, including ImageNet subsets, CIFAR-10, and the MedMNIST histopathological dataset. Our approach achieves state-of-the-art cross architecture performance across a range of backbone architectures, demonstrating its generality and effectiveness.
Video Dataset Condensation with Diffusion Models
May 10, 2025Abstract:In recent years, the rapid expansion of dataset sizes and the increasing complexity of deep learning models have significantly escalated the demand for computational resources, both for data storage and model training. Dataset distillation has emerged as a promising solution to address this challenge by generating a compact synthetic dataset that retains the essential information from a large real dataset. However, existing methods often suffer from limited performance and poor data quality, particularly in the video domain. In this paper, we focus on video dataset distillation by employing a video diffusion model to generate high-quality synthetic videos. To enhance representativeness, we introduce Video Spatio-Temporal U-Net (VST-UNet), a model designed to select a diverse and informative subset of videos that effectively captures the characteristics of the original dataset. To further optimize computational efficiency, we explore a training-free clustering algorithm, Temporal-Aware Cluster-based Distillation (TAC-DT), to select representative videos without requiring additional training overhead. We validate the effectiveness of our approach through extensive experiments on four benchmark datasets, demonstrating performance improvements of up to \(10.61\%\) over the state-of-the-art. Our method consistently outperforms existing approaches across all datasets, establishing a new benchmark for video dataset distillation.
Image Generation Diversity Issues and How to Tame Them
Nov 25, 2024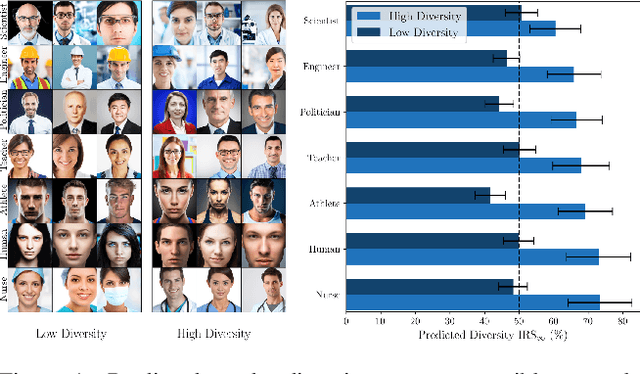
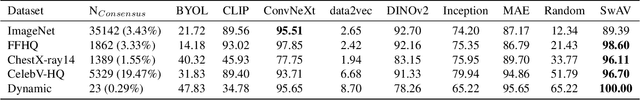
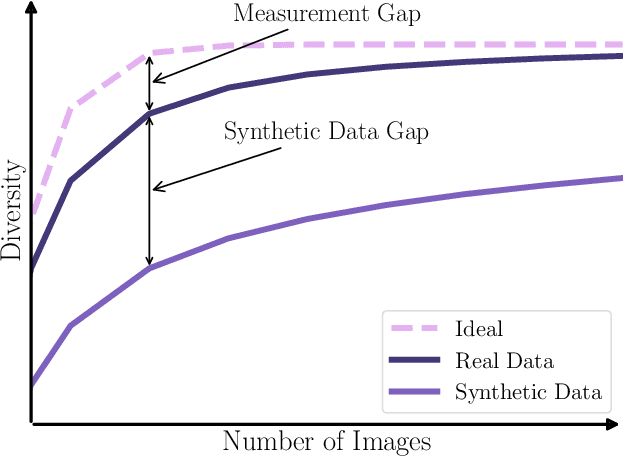
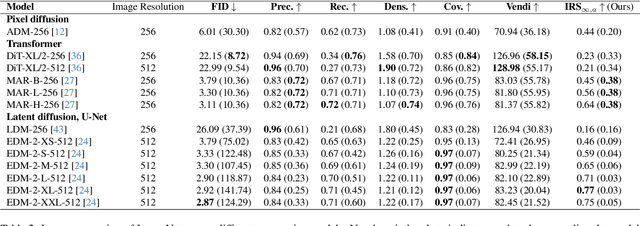
Abstract:Generative methods now produce outputs nearly indistinguishable from real data but often fail to fully capture the data distribution. Unlike quality issues, diversity limitations in generative models are hard to detect visually, requiring specific metrics for assessment. In this paper, we draw attention to the current lack of diversity in generative models and the inability of common metrics to measure this. We achieve this by framing diversity as an image retrieval problem, where we measure how many real images can be retrieved using synthetic data as queries. This yields the Image Retrieval Score (IRS), an interpretable, hyperparameter-free metric that quantifies the diversity of a generative model's output. IRS requires only a subset of synthetic samples and provides a statistical measure of confidence. Our experiments indicate that current feature extractors commonly used in generative model assessment are inadequate for evaluating diversity effectively. Consequently, we perform an extensive search for the best feature extractors to assess diversity. Evaluation reveals that current diffusion models converge to limited subsets of the real distribution, with no current state-of-the-art models superpassing 77% of the diversity of the training data. To address this limitation, we introduce Diversity-Aware Diffusion Models (DiADM), a novel approach that improves diversity of unconditional diffusion models without loss of image quality. We do this by disentangling diversity from image quality by using a diversity aware module that uses pseudo-unconditional features as input. We provide a Python package offering unified feature extraction and metric computation to further facilitate the evaluation of generative models https://github.com/MischaD/beyondfid.
JVID: Joint Video-Image Diffusion for Visual-Quality and Temporal-Consistency in Video Generation
Sep 21, 2024



Abstract:We introduce the Joint Video-Image Diffusion model (JVID), a novel approach to generating high-quality and temporally coherent videos. We achieve this by integrating two diffusion models: a Latent Image Diffusion Model (LIDM) trained on images and a Latent Video Diffusion Model (LVDM) trained on video data. Our method combines these models in the reverse diffusion process, where the LIDM enhances image quality and the LVDM ensures temporal consistency. This unique combination allows us to effectively handle the complex spatio-temporal dynamics in video generation. Our results demonstrate quantitative and qualitative improvements in producing realistic and coherent videos.
Data-Efficient Generation for Dataset Distillation
Sep 05, 2024



Abstract:While deep learning techniques have proven successful in image-related tasks, the exponentially increased data storage and computation costs become a significant challenge. Dataset distillation addresses these challenges by synthesizing only a few images for each class that encapsulate all essential information. Most current methods focus on matching. The problems lie in the synthetic images not being human-readable and the dataset performance being insufficient for downstream learning tasks. Moreover, the distillation time can quickly get out of bounds when the number of synthetic images per class increases even slightly. To address this, we train a class conditional latent diffusion model capable of generating realistic synthetic images with labels. The sampling time can be reduced to several tens of images per seconds. We demonstrate that models can be effectively trained using only a small set of synthetic images and evaluated on a large real test set. Our approach achieved rank \(1\) in The First Dataset Distillation Challenge at ECCV 2024 on the CIFAR100 and TinyImageNet datasets.
URCDM: Ultra-Resolution Image Synthesis in Histopathology
Jul 18, 2024Abstract:Diagnosing medical conditions from histopathology data requires a thorough analysis across the various resolutions of Whole Slide Images (WSI). However, existing generative methods fail to consistently represent the hierarchical structure of WSIs due to a focus on high-fidelity patches. To tackle this, we propose Ultra-Resolution Cascaded Diffusion Models (URCDMs) which are capable of synthesising entire histopathology images at high resolutions whilst authentically capturing the details of both the underlying anatomy and pathology at all magnification levels. We evaluate our method on three separate datasets, consisting of brain, breast and kidney tissue, and surpass existing state-of-the-art multi-resolution models. Furthermore, an expert evaluation study was conducted, demonstrating that URCDMs consistently generate outputs across various resolutions that trained evaluators cannot distinguish from real images. All code and additional examples can be found on GitHub.
Stability and Generalizability in SDE Diffusion Models with Measure-Preserving Dynamics
Jun 19, 2024Abstract:Inverse problems describe the process of estimating the causal factors from a set of measurements or data. Mapping of often incomplete or degraded data to parameters is ill-posed, thus data-driven iterative solutions are required, for example when reconstructing clean images from poor signals. Diffusion models have shown promise as potent generative tools for solving inverse problems due to their superior reconstruction quality and their compatibility with iterative solvers. However, most existing approaches are limited to linear inverse problems represented as Stochastic Differential Equations (SDEs). This simplification falls short of addressing the challenging nature of real-world problems, leading to amplified cumulative errors and biases. We provide an explanation for this gap through the lens of measure-preserving dynamics of Random Dynamical Systems (RDS) with which we analyse Temporal Distribution Discrepancy and thus introduce a theoretical framework based on RDS for SDE diffusion models. We uncover several strategies that inherently enhance the stability and generalizability of diffusion models for inverse problems and introduce a novel score-based diffusion framework, the \textbf{D}ynamics-aware S\textbf{D}E \textbf{D}iffusion \textbf{G}enerative \textbf{M}odel (D$^3$GM). The \textit{Measure-preserving property} can return the degraded measurement to the original state despite complex degradation with the RDS concept of \textit{stability}. Our extensive experimental results corroborate the effectiveness of D$^3$GM across multiple benchmarks including a prominent application for inverse problems, magnetic resonance imaging. Code and data will be publicly available.
Ultra-Resolution Cascaded Diffusion Model for Gigapixel Image Synthesis in Histopathology
Dec 02, 2023


Abstract:Diagnoses from histopathology images rely on information from both high and low resolutions of Whole Slide Images. Ultra-Resolution Cascaded Diffusion Models (URCDMs) allow for the synthesis of high-resolution images that are realistic at all magnification levels, focusing not only on fidelity but also on long-distance spatial coherency. Our model beats existing methods, improving the pFID-50k [2] score by 110.63 to 39.52 pFID-50k. Additionally, a human expert evaluation study was performed, reaching a weighted Mean Absolute Error (MAE) of 0.11 for the Lower Resolution Diffusion Models and a weighted MAE of 0.22 for the URCDM.
Realistic Data Enrichment for Robust Image Segmentation in Histopathology
Apr 19, 2023


Abstract:Poor performance of quantitative analysis in histopathological Whole Slide Images (WSI) has been a significant obstacle in clinical practice. Annotating large-scale WSIs manually is a demanding and time-consuming task, unlikely to yield the expected results when used for fully supervised learning systems. Rarely observed disease patterns and large differences in object scales are difficult to model through conventional patient intake. Prior methods either fall back to direct disease classification, which only requires learning a few factors per image, or report on average image segmentation performance, which is highly biased towards majority observations. Geometric image augmentation is commonly used to improve robustness for average case predictions and to enrich limited datasets. So far no method provided sampling of a realistic posterior distribution to improve stability, e.g. for the segmentation of imbalanced objects within images. Therefore, we propose a new approach, based on diffusion models, which can enrich an imbalanced dataset with plausible examples from underrepresented groups by conditioning on segmentation maps. Our method can simply expand limited clinical datasets making them suitable to train machine learning pipelines, and provides an interpretable and human-controllable way of generating histopathology images that are indistinguishable from real ones to human experts. We validate our findings on two datasets, one from the public domain and one from a Kidney Transplant study.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge