Rossella Arcucci
Imperial College London, United Kingdom, Data Science Institute, Imperial College London, United Kingdom
A Hybrid Machine Learning Framework for Improved Short-Term Peak-Flow Forecasting
Jan 14, 2026Abstract:Reliable river flow forecasting is an essential component of flood risk management and early warning systems. It enables improved emergency response coordination and is critical for protecting infrastructure, communities, and ecosystems from extreme hydrological events. Process-based hydrological models and purely data-driven approaches often underperform during extreme events, particularly in forecasting peak flows. To address this limitation, this study introduces a hybrid forecasting framework that couples Extreme Gradient Boosting (XGBoost) and Random Forest (RF). XGBoost is employed for continuous streamflow forecasting, while RF is specifically trained for peak-flow prediction, and the two outputs are combined into an enhanced forecast. The approach is implemented across 857 catchments of the LamaH-CE dataset, using rainfall and discharge observations at 6-hour resolution. Results demonstrate consistently high skill, with 71% of catchments achieving a Kling-Gupta Efficiency (KGE) greater than 0.90. Peak-flow detection reaches 87%, with a false-alarm rate of 13%. Compared to the European Flood Awareness System (EFAS), the framework achieves lower peak-magnitude errors, fewer false alarms, and improved streamflow and peak-flow forecasting accuracy. The proposed framework is computationally lightweight, scalable, and easily transferable across watersheds, with training times of only seconds on standard CPUs. These findings highlight the potential of integrating hydrological understanding with efficient machine learning to improve the accuracy and reliability of operational flood forecasting, and outline future directions for hybrid hydrological-machine learning model development.
Knowledge to Sight: Reasoning over Visual Attributes via Knowledge Decomposition for Abnormality Grounding
Aug 06, 2025Abstract:In this work, we address the problem of grounding abnormalities in medical images, where the goal is to localize clinical findings based on textual descriptions. While generalist Vision-Language Models (VLMs) excel in natural grounding tasks, they often struggle in the medical domain due to rare, compositional, and domain-specific terms that are poorly aligned with visual patterns. Specialized medical VLMs address this challenge via large-scale domain pretraining, but at the cost of substantial annotation and computational resources. To overcome these limitations, we propose \textbf{Knowledge to Sight (K2Sight)}, a framework that introduces structured semantic supervision by decomposing clinical concepts into interpretable visual attributes, such as shape, density, and anatomical location. These attributes are distilled from domain ontologies and encoded into concise instruction-style prompts, which guide region-text alignment during training. Unlike conventional report-level supervision, our approach explicitly bridges domain knowledge and spatial structure, enabling data-efficient training of compact models. We train compact models with 0.23B and 2B parameters using only 1.5\% of the data required by state-of-the-art medical VLMs. Despite their small size and limited training data, these models achieve performance on par with or better than 7B+ medical VLMs, with up to 9.82\% improvement in $mAP_{50}$. Code and models: \href{https://lijunrio.github.io/K2Sight/}{\textcolor{SOTAPink}{https://lijunrio.github.io/K2Sight/}}.
BOTM: Echocardiography Segmentation via Bi-directional Optimal Token Matching
May 23, 2025Abstract:Existed echocardiography segmentation methods often suffer from anatomical inconsistency challenge caused by shape variation, partial observation and region ambiguity with similar intensity across 2D echocardiographic sequences, resulting in false positive segmentation with anatomical defeated structures in challenging low signal-to-noise ratio conditions. To provide a strong anatomical guarantee across different echocardiographic frames, we propose a novel segmentation framework named BOTM (Bi-directional Optimal Token Matching) that performs echocardiography segmentation and optimal anatomy transportation simultaneously. Given paired echocardiographic images, BOTM learns to match two sets of discrete image tokens by finding optimal correspondences from a novel anatomical transportation perspective. We further extend the token matching into a bi-directional cross-transport attention proxy to regulate the preserved anatomical consistency within the cardiac cyclic deformation in temporal domain. Extensive experimental results show that BOTM can generate stable and accurate segmentation outcomes (e.g. -1.917 HD on CAMUS2H LV, +1.9% Dice on TED), and provide a better matching interpretation with anatomical consistency guarantee.
Beyond Distillation: Pushing the Limits of Medical LLM Reasoning with Minimalist Rule-Based RL
May 23, 2025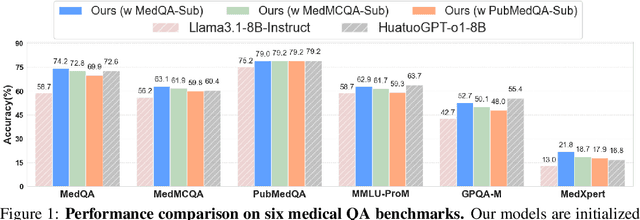
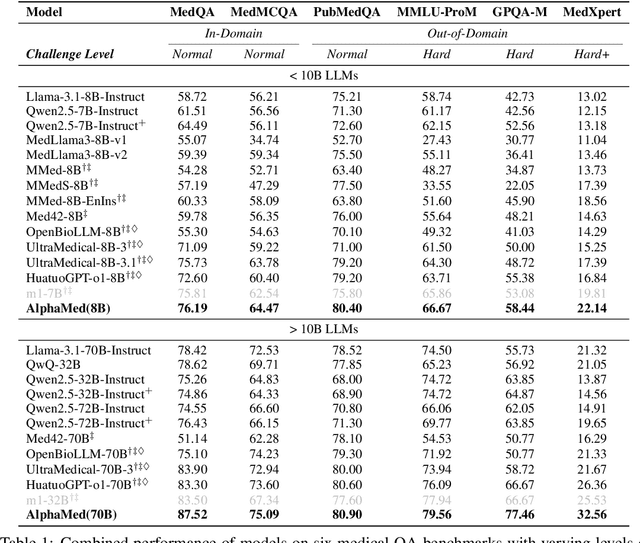
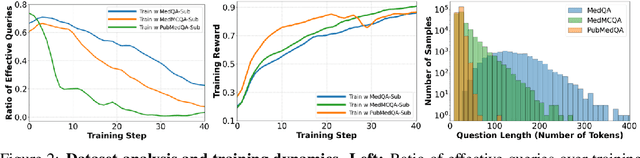
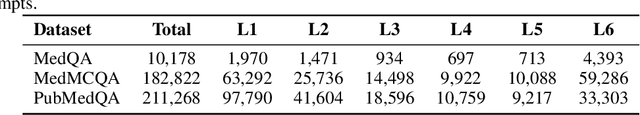
Abstract:Improving performance on complex tasks and enabling interpretable decision making in large language models (LLMs), especially for clinical applications, requires effective reasoning. Yet this remains challenging without supervised fine-tuning (SFT) on costly chain-of-thought (CoT) data distilled from closed-source models (e.g., GPT-4o). In this work, we present AlphaMed, the first medical LLM to show that reasoning capability can emerge purely through reinforcement learning (RL), using minimalist rule-based rewards on public multiple-choice QA datasets, without relying on SFT or distilled CoT data. AlphaMed achieves state-of-the-art results on six medical QA benchmarks, outperforming models trained with conventional SFT+RL pipelines. On challenging benchmarks (e.g., MedXpert), AlphaMed even surpasses larger or closed-source models such as DeepSeek-V3-671B and Claude-3.5-Sonnet. To understand the factors behind this success, we conduct a comprehensive data-centric analysis guided by three questions: (i) Can minimalist rule-based RL incentivize reasoning without distilled CoT supervision? (ii) How do dataset quantity and diversity impact reasoning? (iii) How does question difficulty shape the emergence and generalization of reasoning? Our findings show that dataset informativeness is a key driver of reasoning performance, and that minimalist RL on informative, multiple-choice QA data is effective at inducing reasoning without CoT supervision. We also observe divergent trends across benchmarks, underscoring limitations in current evaluation and the need for more challenging, reasoning-oriented medical QA benchmarks.
Enhancing Abnormality Grounding for Vision Language Models with Knowledge Descriptions
Mar 05, 2025Abstract:Visual Language Models (VLMs) have demonstrated impressive capabilities in visual grounding tasks. However, their effectiveness in the medical domain, particularly for abnormality detection and localization within medical images, remains underexplored. A major challenge is the complex and abstract nature of medical terminology, which makes it difficult to directly associate pathological anomaly terms with their corresponding visual features. In this work, we introduce a novel approach to enhance VLM performance in medical abnormality detection and localization by leveraging decomposed medical knowledge. Instead of directly prompting models to recognize specific abnormalities, we focus on breaking down medical concepts into fundamental attributes and common visual patterns. This strategy promotes a stronger alignment between textual descriptions and visual features, improving both the recognition and localization of abnormalities in medical images.We evaluate our method on the 0.23B Florence-2 base model and demonstrate that it achieves comparable performance in abnormality grounding to significantly larger 7B LLaVA-based medical VLMs, despite being trained on only 1.5% of the data used for such models. Experimental results also demonstrate the effectiveness of our approach in both known and previously unseen abnormalities, suggesting its strong generalization capabilities.
SuPreME: A Supervised Pre-training Framework for Multimodal ECG Representation Learning
Feb 27, 2025Abstract:Cardiovascular diseases are a leading cause of death and disability worldwide. Electrocardiogram (ECG) recordings are critical for diagnosing and monitoring cardiac health, but obtaining large-scale annotated ECG datasets is labor-intensive and time-consuming. Recent ECG Self-Supervised Learning (eSSL) methods mitigate this by learning features without extensive labels but fail to capture fine-grained clinical semantics and require extensive task-specific fine-tuning. To address these challenges, we propose $\textbf{SuPreME}$, a $\textbf{Su}$pervised $\textbf{Pre}$-training framework for $\textbf{M}$ultimodal $\textbf{E}$CG representation learning. SuPreME applies Large Language Models (LLMs) to extract structured clinical entities from free-text ECG reports, filter out noise and irrelevant content, enhance clinical representation learning, and build a high-quality, fine-grained labeled dataset. By using text-based cardiac queries instead of traditional categorical labels, SuPreME enables zero-shot classification of unseen diseases without additional fine-tuning. We evaluate SuPreME on six downstream datasets covering 127 cardiac conditions, achieving superior zero-shot AUC performance over state-of-the-art eSSL and multimodal methods by over 1.96\%. Results demonstrate the effectiveness of SuPreME in leveraging structured, clinically relevant knowledge for high-quality ECG representations. All code and data will be released upon acceptance.
Knowledge-enhanced Multimodal ECG Representation Learning with Arbitrary-Lead Inputs
Feb 25, 2025



Abstract:Recent advances in multimodal ECG representation learning center on aligning ECG signals with paired free-text reports. However, suboptimal alignment persists due to the complexity of medical language and the reliance on a full 12-lead setup, which is often unavailable in under-resourced settings. To tackle these issues, we propose **K-MERL**, a knowledge-enhanced multimodal ECG representation learning framework. **K-MERL** leverages large language models to extract structured knowledge from free-text reports and employs a lead-aware ECG encoder with dynamic lead masking to accommodate arbitrary lead inputs. Evaluations on six external ECG datasets show that **K-MERL** achieves state-of-the-art performance in zero-shot classification and linear probing tasks, while delivering an average **16%** AUC improvement over existing methods in partial-lead zero-shot classification.
Machine learning for modelling unstructured grid data in computational physics: a review
Feb 13, 2025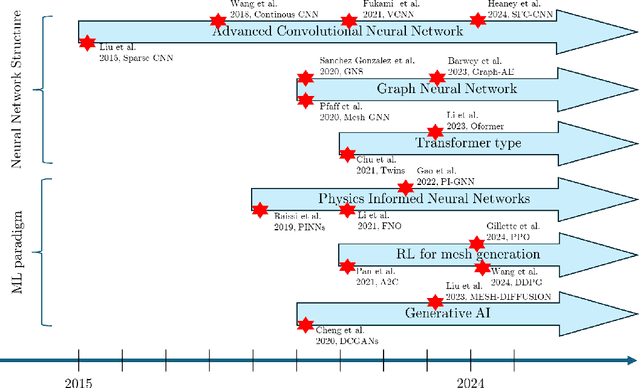
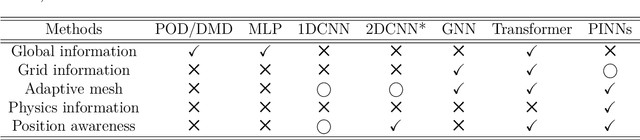
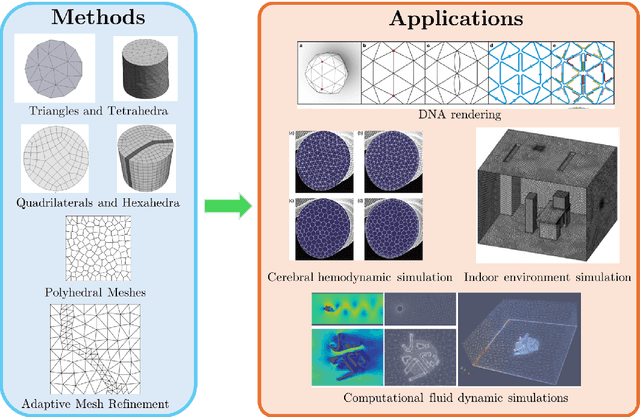
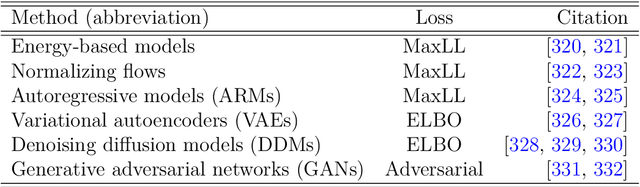
Abstract:Unstructured grid data are essential for modelling complex geometries and dynamics in computational physics. Yet, their inherent irregularity presents significant challenges for conventional machine learning (ML) techniques. This paper provides a comprehensive review of advanced ML methodologies designed to handle unstructured grid data in high-dimensional dynamical systems. Key approaches discussed include graph neural networks, transformer models with spatial attention mechanisms, interpolation-integrated ML methods, and meshless techniques such as physics-informed neural networks. These methodologies have proven effective across diverse fields, including fluid dynamics and environmental simulations. This review is intended as a guidebook for computational scientists seeking to apply ML approaches to unstructured grid data in their domains, as well as for ML researchers looking to address challenges in computational physics. It places special focus on how ML methods can overcome the inherent limitations of traditional numerical techniques and, conversely, how insights from computational physics can inform ML development. To support benchmarking, this review also provides a summary of open-access datasets of unstructured grid data in computational physics. Finally, emerging directions such as generative models with unstructured data, reinforcement learning for mesh generation, and hybrid physics-data-driven paradigms are discussed to inspire future advancements in this evolving field.
DYffCast: Regional Precipitation Nowcasting Using IMERG Satellite Data. A case study over South America
Dec 02, 2024
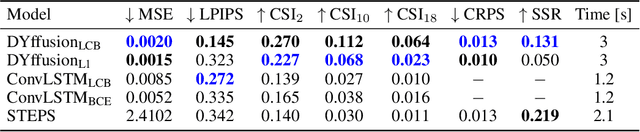
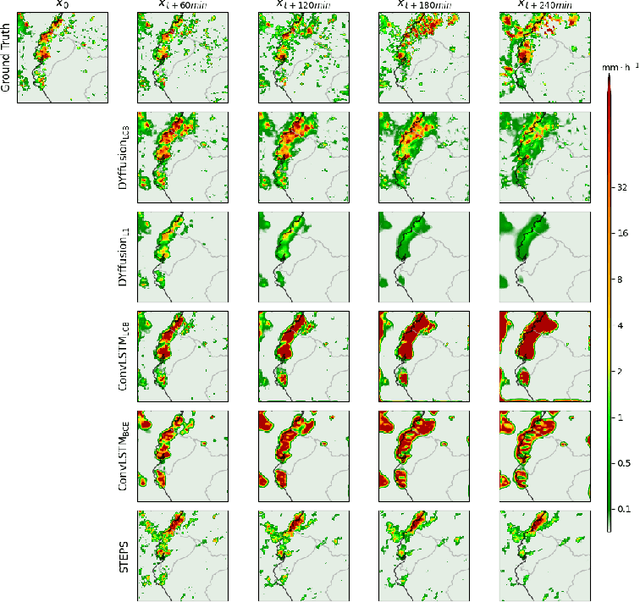
Abstract:Climate change is increasing the frequency of extreme precipitation events, making weather disasters such as flooding and landslides more likely. The ability to accurately nowcast precipitation is therefore becoming more critical for safeguarding society by providing immediate, accurate information to decision makers. Motivated by the recent success of generative models at precipitation nowcasting, this paper: extends the DYffusion framework to this task and evaluates its performance at forecasting IMERG satellite precipitation data up to a 4-hour horizon; modifies the DYffusion framework to improve its ability to model rainfall data; and introduces a novel loss function that combines MSE, MAE and the LPIPS perceptual score. In a quantitative evaluation of forecasts up to a 4-hour horizon, the modified DYffusion framework trained with the novel loss outperforms four competitor models. It has the highest CSI scores for weak, moderate, and heavy rain thresholds and retains an LPIPS score $<$ 0.2 for the entire roll-out, degrading the least as lead-time increases. The proposed nowcasting model demonstrates visually stable and sharp forecasts up to a 2-hour horizon on a heavy rain case study. Code is available at https://github.com/Dseal95/DYffcast.
Fire-Image-DenseNet (FIDN) for predicting wildfire burnt area using remote sensing data
Dec 02, 2024



Abstract:Predicting the extent of massive wildfires once ignited is essential to reduce the subsequent socioeconomic losses and environmental damage, but challenging because of the complexity of fire behaviour. Existing physics-based models are limited in predicting large or long-duration wildfire events. Here, we develop a deep-learning-based predictive model, Fire-Image-DenseNet (FIDN), that uses spatial features derived from both near real-time and reanalysis data on the environmental and meteorological drivers of wildfire. We trained and tested this model using more than 300 individual wildfires that occurred between 2012 and 2019 in the western US. In contrast to existing models, the performance of FIDN does not degrade with fire size or duration. Furthermore, it predicts final burnt area accurately even in very heterogeneous landscapes in terms of fuel density and flammability. The FIDN model showed higher accuracy, with a mean squared error (MSE) about 82% and 67% lower than those of the predictive models based on cellular automata (CA) and the minimum travel time (MTT) approaches, respectively. Its structural similarity index measure (SSIM) averages 97%, outperforming the CA and FlamMap MTT models by 6% and 2%, respectively. Additionally, FIDN is approximately three orders of magnitude faster than both CA and MTT models. The enhanced computational efficiency and accuracy advancements offer vital insights for strategic planning and resource allocation for firefighting operations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge