Fariba Yousefi
Segment Anyword: Mask Prompt Inversion for Open-Set Grounded Segmentation
May 23, 2025Abstract:Open-set image segmentation poses a significant challenge because existing methods often demand extensive training or fine-tuning and generally struggle to segment unified objects consistently across diverse text reference expressions. Motivated by this, we propose Segment Anyword, a novel training-free visual concept prompt learning approach for open-set language grounded segmentation that relies on token-level cross-attention maps from a frozen diffusion model to produce segmentation surrogates or mask prompts, which are then refined into targeted object masks. Initial prompts typically lack coherence and consistency as the complexity of the image-text increases, resulting in suboptimal mask fragments. To tackle this issue, we further introduce a novel linguistic-guided visual prompt regularization that binds and clusters visual prompts based on sentence dependency and syntactic structural information, enabling the extraction of robust, noise-tolerant mask prompts, and significant improvements in segmentation accuracy. The proposed approach is effective, generalizes across different open-set segmentation tasks, and achieves state-of-the-art results of 52.5 (+6.8 relative) mIoU on Pascal Context 59, 67.73 (+25.73 relative) cIoU on gRefCOCO, and 67.4 (+1.1 relative to fine-tuned methods) mIoU on GranDf, which is the most complex open-set grounded segmentation task in the field.
MiceBoneChallenge: Micro-CT public dataset and six solutions for automatic growth plate detection in micro-CT mice bone scans
Nov 26, 2024



Abstract:Detecting and quantifying bone changes in micro-CT scans of rodents is a common task in preclinical drug development studies. However, this task is manual, time-consuming and subject to inter- and intra-observer variability. In 2024, Anonymous Company organized an internal challenge to develop models for automatic bone quantification. We prepared and annotated a high-quality dataset of 3D $\mu$CT bone scans from $83$ mice. The challenge attracted over $80$ AI scientists from around the globe who formed $23$ teams. The participants were tasked with developing a solution to identify the plane where the bone growth happens, which is essential for fully automatic segmentation of trabecular bone. As a result, six computer vision solutions were developed that can accurately identify the location of the growth plate plane. The solutions achieved the mean absolute error of $1.91\pm0.87$ planes from the ground truth on the test set, an accuracy level acceptable for practical use by a radiologist. The annotated 3D scans dataset along with the six solutions and source code, is being made public, providing researchers with opportunities to develop and benchmark their own approaches. The code, trained models, and the data will be shared.
Lung tumor segmentation in MRI mice scans using 3D nnU-Net with minimum annotations
Nov 01, 2024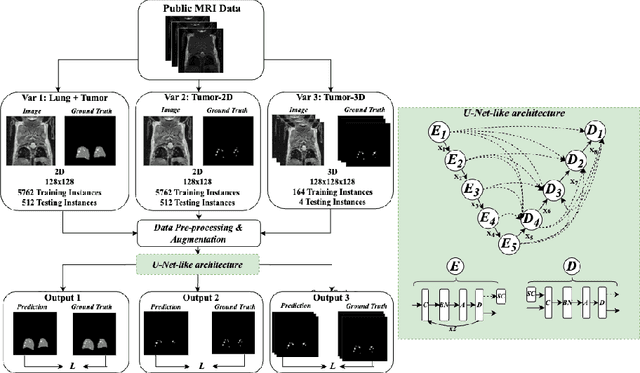
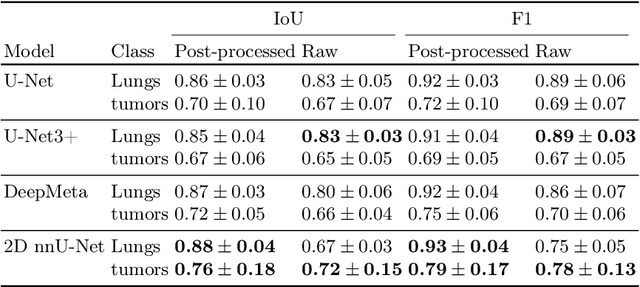
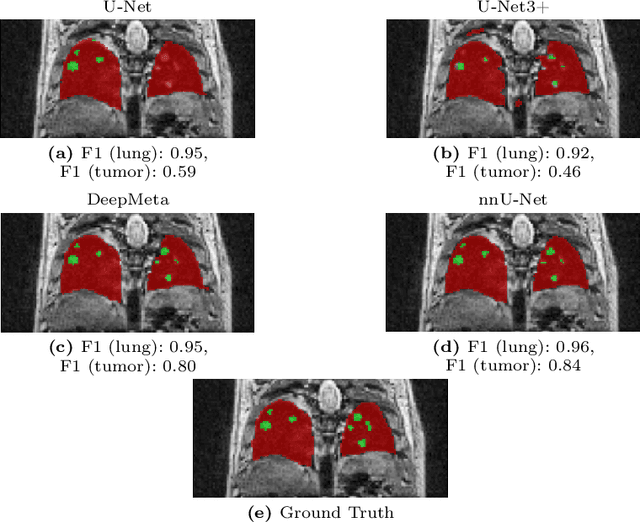
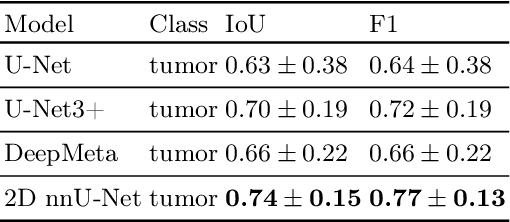
Abstract:In drug discovery, accurate lung tumor segmentation is an important step for assessing tumor size and its progression using \textit{in-vivo} imaging such as MRI. While deep learning models have been developed to automate this process, the focus has predominantly been on human subjects, neglecting the pivotal role of animal models in pre-clinical drug development. In this work, we focus on optimizing lung tumor segmentation in mice. First, we demonstrate that the nnU-Net model outperforms the U-Net, U-Net3+, and DeepMeta models. Most importantly, we achieve better results with nnU-Net 3D models than 2D models, indicating the importance of spatial context for segmentation tasks in MRI mice scans. This study demonstrates the importance of 3D input over 2D input images for lung tumor segmentation in MRI scans. Finally, we outperform the prior state-of-the-art approach that involves the combined segmentation of lungs and tumors within the lungs. Our work achieves comparable results using only lung tumor annotations requiring fewer annotations, saving time and annotation efforts. This work\footnote{\url{https://anonymous.4open.science/r/lung-tumour-mice-mri-64BB}} is an important step in automating pre-clinical animal studies to quantify the efficacy of experimental drugs, particularly in assessing tumor changes.
Can Medical Vision-Language Pre-training Succeed with Purely Synthetic Data?
Oct 17, 2024



Abstract:Medical Vision-Language Pre-training (MedVLP) has made significant progress in enabling zero-shot tasks for medical image understanding. However, training MedVLP models typically requires large-scale datasets with paired, high-quality image-text data, which are scarce in the medical domain. Recent advancements in Large Language Models (LLMs) and diffusion models have made it possible to generate large-scale synthetic image-text pairs. This raises the question: *Can MedVLP succeed using purely synthetic data?* To address this, we use off-the-shelf generative models to create synthetic radiology reports and paired Chest X-ray (CXR) images, and propose an automated pipeline to build a diverse, high-quality synthetic dataset, enabling a rigorous study that isolates model and training settings, focusing entirely from the data perspective. Our results show that MedVLP models trained *exclusively on synthetic data* outperform those trained on real data by **3.8%** in averaged AUC on zero-shot classification. Moreover, using a combination of synthetic and real data leads to a further improvement of **9.07%**. Additionally, MedVLP models trained on synthetic or mixed data consistently outperform those trained on real data in zero-shot grounding, as well as in fine-tuned classification and segmentation tasks. Our analysis suggests MedVLP trained on well-designed synthetic data can outperform models trained on real datasets, which may be limited by low-quality samples and long-tailed distributions.
Convening during COVID-19: Lessons learnt from organizing virtual workshops in 2020
Nov 28, 2020Abstract:This report is an account of the authors' experiences as organizers of WiML's "Un-Workshop" event at ICML 2020. Un-workshops focus on participant-driven structured discussions on a pre-selected topic. For clarity, this event was different from the "WiML Workshop", which is usually co-located with NeurIPS. In this manuscript, organizers, share their experiences with the hope that it will help future organizers to host a successful virtual event under similar conditions. Women in Machine Learning (WiML)'s mission is creating connections within a small community of women working in machine learning, in order to encourage mentorship, networking, and interchange of ideas and increase the impact of women in the community.
Multi-task Learning for Aggregated Data using Gaussian Processes
Jun 22, 2019



Abstract:Aggregated data is commonplace in areas such as epidemiology and demography. For example, census data for a population is usually given as averages defined over time periods or spatial resolutions (city, region or countries). In this paper, we present a novel multi-task learning model based on Gaussian processes for joint learning of variables that have been aggregated at different input scales. Our model represents each task as the linear combination of the realizations of latent processes that are integrated at a different scale per task. We are then able to compute the cross-covariance between the different tasks either analytically or numerically. We also allow each task to have a potentially different likelihood model and provide a variational lower bound that can be optimised in a stochastic fashion making our model suitable for larger datasets. We show examples of the model in a synthetic example, a fertility dataset and an air pollution prediction application.
Unsupervised Learning with Imbalanced Data via Structure Consolidation Latent Variable Model
Jun 30, 2016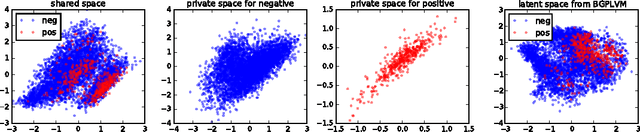
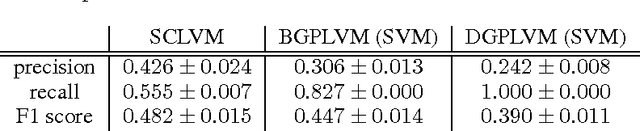
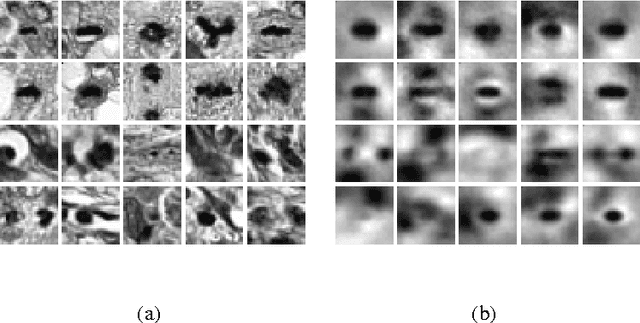
Abstract:Unsupervised learning on imbalanced data is challenging because, when given imbalanced data, current model is often dominated by the major category and ignores the categories with small amount of data. We develop a latent variable model that can cope with imbalanced data by dividing the latent space into a shared space and a private space. Based on Gaussian Process Latent Variable Models, we propose a new kernel formulation that enables the separation of latent space and derives an efficient variational inference method. The performance of our model is demonstrated with an imbalanced medical image dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge