Caroline Weis
Genetic prediction of quantitative traits: a machine learner's guide focused on height
Oct 06, 2023
Abstract:Machine learning and deep learning have been celebrating many successes in the application to biological problems, especially in the domain of protein folding. Another equally complex and important question has received relatively little attention by the machine learning community, namely the one of prediction of complex traits from genetics. Tackling this problem requires in-depth knowledge of the related genetics literature and awareness of various subtleties associated with genetic data. In this guide, we provide an overview for the machine learning community on current state of the art models and associated subtleties which need to be taken into consideration when developing new models for phenotype prediction. We use height as an example of a continuous-valued phenotype and provide an introduction to benchmark datasets, confounders, feature selection, and common metrics.
Convening during COVID-19: Lessons learnt from organizing virtual workshops in 2020
Nov 28, 2020Abstract:This report is an account of the authors' experiences as organizers of WiML's "Un-Workshop" event at ICML 2020. Un-workshops focus on participant-driven structured discussions on a pre-selected topic. For clarity, this event was different from the "WiML Workshop", which is usually co-located with NeurIPS. In this manuscript, organizers, share their experiences with the hope that it will help future organizers to host a successful virtual event under similar conditions. Women in Machine Learning (WiML)'s mission is creating connections within a small community of women working in machine learning, in order to encourage mentorship, networking, and interchange of ideas and increase the impact of women in the community.
Topological Data Analysis of copy number alterations in cancer
Nov 22, 2020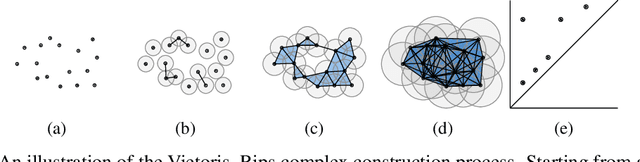



Abstract:Identifying subgroups and properties of cancer biopsy samples is a crucial step towards obtaining precise diagnoses and being able to perform personalized treatment of cancer patients. Recent data collections provide a comprehensive characterization of cancer cell data, including genetic data on copy number alterations (CNAs). We explore the potential to capture information contained in cancer genomic information using a novel topology-based approach that encodes each cancer sample as a persistence diagram of topological features, i.e., high-dimensional voids represented in the data. We find that this technique has the potential to extract meaningful low-dimensional representations in cancer somatic genetic data and demonstrate the viability of some applications on finding substructures in cancer data as well as comparing similarity of cancer types.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge