Mandana Samiei
Language Agents Mirror Human Causal Reasoning Biases. How Can We Help Them Think Like Scientists?
May 14, 2025Abstract:Language model (LM) agents are increasingly used as autonomous decision-makers who need to actively gather information to guide their decisions. A crucial cognitive skill for such agents is the efficient exploration and understanding of the causal structure of the world -- key to robust, scientifically grounded reasoning. Yet, it remains unclear whether LMs possess this capability or exhibit systematic biases leading to erroneous conclusions. In this work, we examine LMs' ability to explore and infer causal relationships, using the well-established "Blicket Test" paradigm from developmental psychology. We find that LMs reliably infer the common, intuitive disjunctive causal relationships but systematically struggle with the unusual, yet equally (or sometimes even more) evidenced conjunctive ones. This "disjunctive bias" persists across model families, sizes, and prompting strategies, and performance further declines as task complexity increases. Interestingly, an analogous bias appears in human adults, suggesting that LMs may have inherited deep-seated reasoning heuristics from their training data. To this end, we quantify similarities between LMs and humans, finding that LMs exhibit adult-like inference profiles (but not children-like). Finally, we propose a test-time sampling method which explicitly samples and eliminates hypotheses about causal relationships from the LM. This scalable approach significantly reduces the disjunctive bias and moves LMs closer to the goal of scientific, causally rigorous reasoning.
Convening during COVID-19: Lessons learnt from organizing virtual workshops in 2020
Nov 28, 2020Abstract:This report is an account of the authors' experiences as organizers of WiML's "Un-Workshop" event at ICML 2020. Un-workshops focus on participant-driven structured discussions on a pre-selected topic. For clarity, this event was different from the "WiML Workshop", which is usually co-located with NeurIPS. In this manuscript, organizers, share their experiences with the hope that it will help future organizers to host a successful virtual event under similar conditions. Women in Machine Learning (WiML)'s mission is creating connections within a small community of women working in machine learning, in order to encourage mentorship, networking, and interchange of ideas and increase the impact of women in the community.
The TCGA Meta-Dataset Clinical Benchmark
Oct 18, 2019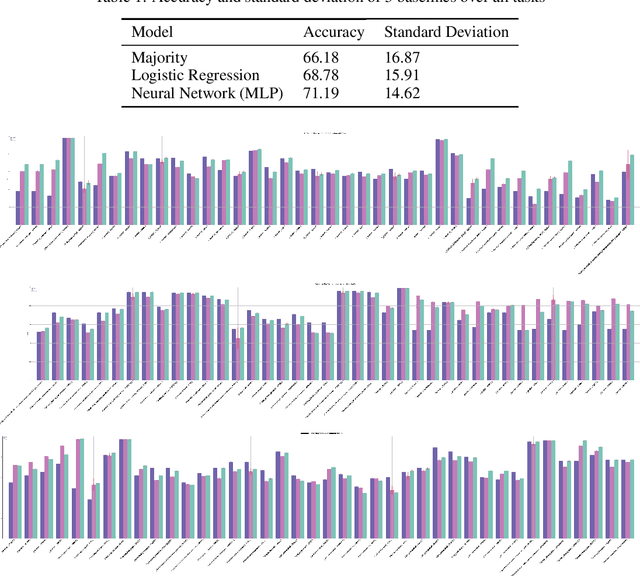
Abstract:Machine learning is bringing a paradigm shift to healthcare by changing the process of disease diagnosis and prognosis in clinics and hospitals. This development equips doctors and medical staff with tools to evaluate their hypotheses and hence make more precise decisions. Although most current research in the literature seeks to develop techniques and methods for predicting one particular clinical outcome, this approach is far from the reality of clinical decision making in which you have to consider several factors simultaneously. In addition, it is difficult to follow the recent progress concretely as there is a lack of consistency in benchmark datasets and task definitions in the field of Genomics. To address the aforementioned issues, we provide a clinical Meta-Dataset derived from the publicly available data hub called The Cancer Genome Atlas Program (TCGA) that contains 174 tasks. We believe those tasks could be good proxy tasks to develop methods which can work on a few samples of gene expression data. Also, learning to predict multiple clinical variables using gene-expression data is an important task due to the variety of phenotypes in clinical problems and lack of samples for some of the rare variables. The defined tasks cover a wide range of clinical problems including predicting tumor tissue site, white cell count, histological type, family history of cancer, gender, and many others which we explain later in the paper. Each task represents an independent dataset. We use regression and neural network baselines for all the tasks using only 150 samples and compare their performance.
Torchmeta: A Meta-Learning library for PyTorch
Sep 14, 2019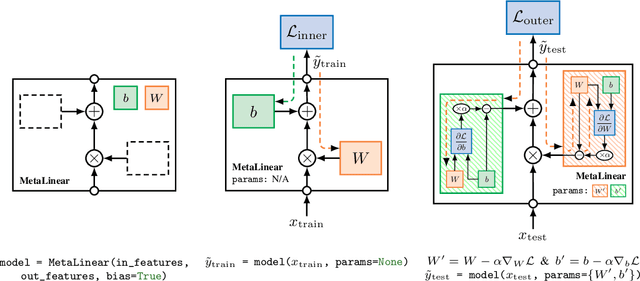
Abstract:The constant introduction of standardized benchmarks in the literature has helped accelerating the recent advances in meta-learning research. They offer a way to get a fair comparison between different algorithms, and the wide range of datasets available allows full control over the complexity of this evaluation. However, for a large majority of code available online, the data pipeline is often specific to one dataset, and testing on another dataset requires significant rework. We introduce Torchmeta, a library built on top of PyTorch that enables seamless and consistent evaluation of meta-learning algorithms on multiple datasets, by providing data-loaders for most of the standard benchmarks in few-shot classification and regression, with a new meta-dataset abstraction. It also features some extensions for PyTorch to simplify the development of models compatible with meta-learning algorithms. The code is available here: https://github.com/tristandeleu/pytorch-meta
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge