Tobias Würfl
2-Step Sparse-View CT Reconstruction with a Domain-Specific Perceptual Network
Dec 08, 2020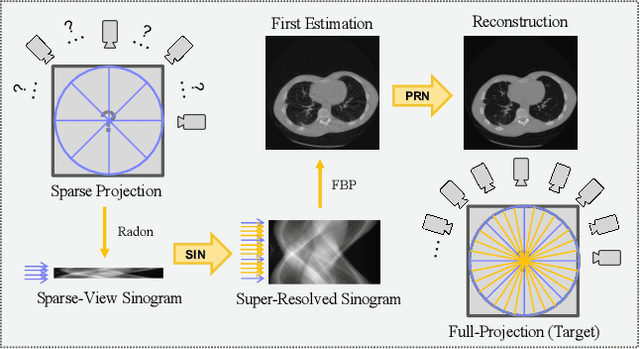
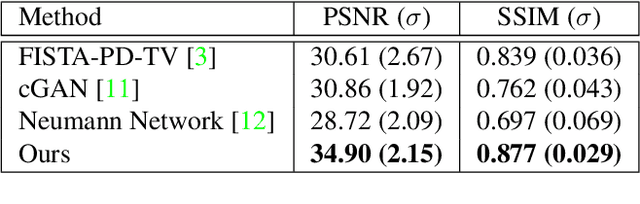
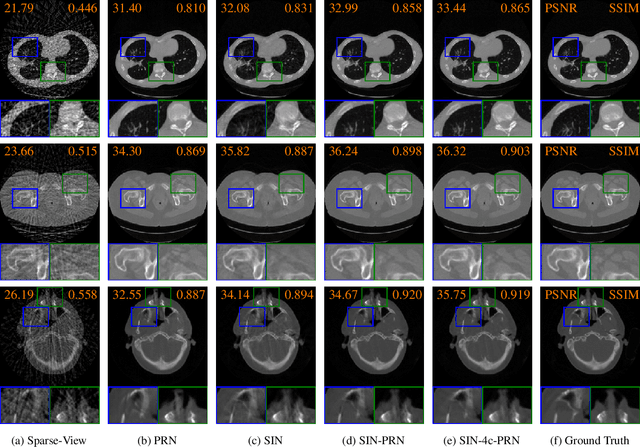
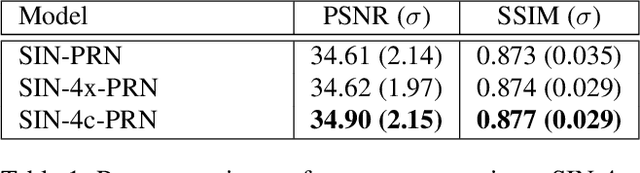
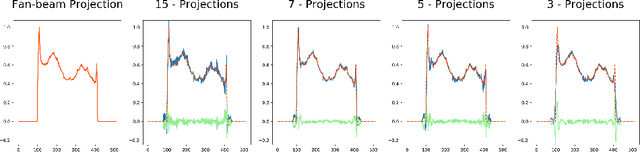
Abstract:Computed tomography is widely used to examine internal structures in a non-destructive manner. To obtain high-quality reconstructions, one typically has to acquire a densely sampled trajectory to avoid angular undersampling. However, many scenarios require a sparse-view measurement leading to streak-artifacts if unaccounted for. Current methods do not make full use of the domain-specific information, and hence fail to provide reliable reconstructions for highly undersampled data. We present a novel framework for sparse-view tomography by decoupling the reconstruction into two steps: First, we overcome its ill-posedness using a super-resolution network, SIN, trained on the sparse projections. The intermediate result allows for a closed-form tomographic reconstruction with preserved details and highly reduced streak-artifacts. Second, a refinement network, PRN, trained on the reconstructions reduces any remaining artifacts. We further propose a light-weight variant of the perceptual-loss that enhances domain-specific information, boosting restoration accuracy. Our experiments demonstrate an improvement over current solutions by 4 dB.
Disassemblable Fieldwork CT Scanner Using a 3D-printed Calibration Phantom
Nov 12, 2020



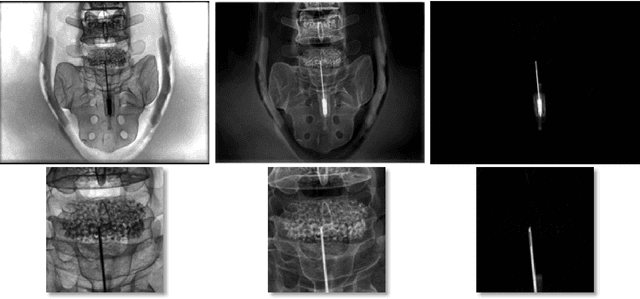
Abstract:The use of computed tomography (CT) imaging has become of increasing interest to academic areas outside of the field of medical imaging and industrial inspection, e.g., to biology and cultural heritage research. The pecularities of these fields, however, sometimes require that objects need to be imaged on-site, e.g., in field-work conditions or in museum collections. Under these circumstances, it is often not possible to use a commercial device and a custom solution is the only viable option. In order to achieve high image quality under adverse conditions, reliable calibration and trajectory reproduction are usually key requirements for any custom CT scanning system. Here, we introduce the construction of a low-cost disassemblable CT scanner that allows calibration even when trajectory reproduction is not possible due to the limitations imposed by the project conditions. Using 3D-printed in-image calibration phantoms, we compute a projection matrix directly from each captured X-ray projection. We describe our method in detail and show successful tomographic reconstructions of several specimen as proof of concept.
* This paper was originally published at the 6th International Conference on Image Formation in X-Ray Computed Tomography (CTmeeting 2020)
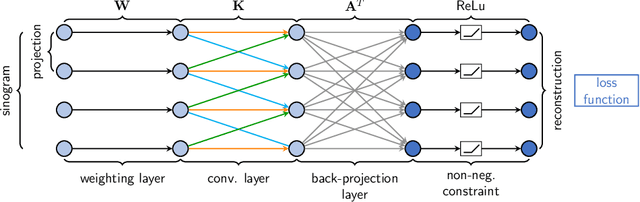
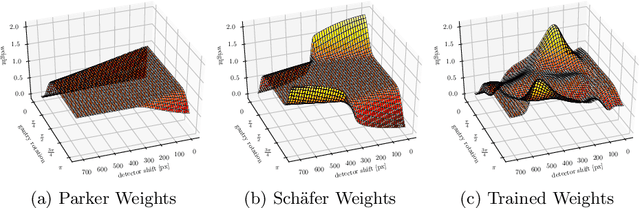
Reconstruction of Voxels with Position- and Angle-Dependent Weightings
Oct 27, 2020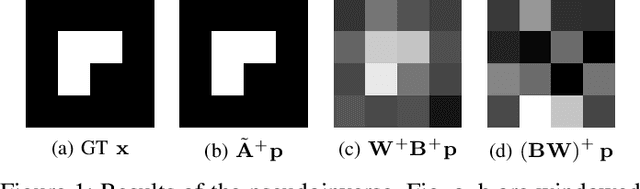
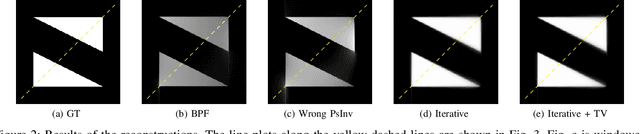
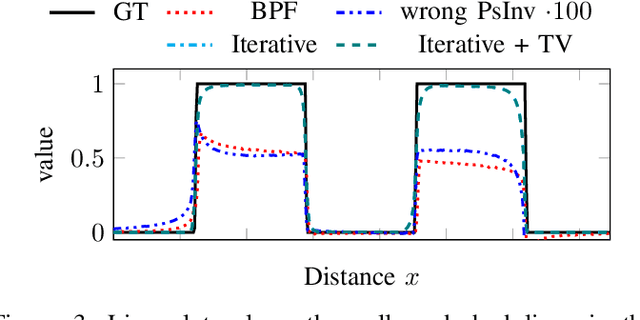
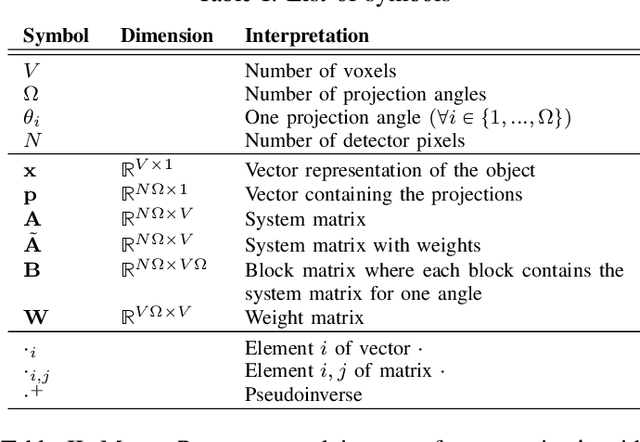
Abstract:The reconstruction problem of voxels with individual weightings can be modeled a position- and angle- dependent function in the forward-projection. This changes the system matrix and prohibits to use standard filtered backprojection. In this work we first formulate this reconstruction problem in terms of a system matrix and weighting part. We compute the pseudoinverse and show that the solution is rank-deficient and hence very ill posed. This is a fundamental limitation for reconstruction. We then derive an iterative solution and experimentally show its uperiority to any closed-form solution.
Deep Learning-based Pipeline for Module Power Prediction from EL Measurements
Sep 30, 2020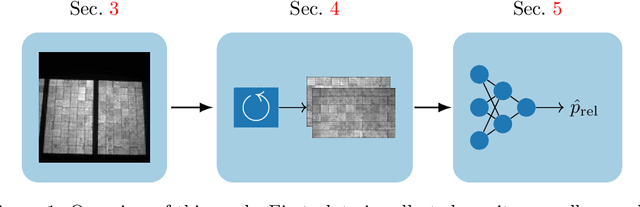

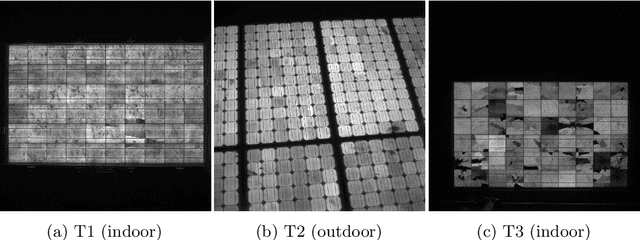
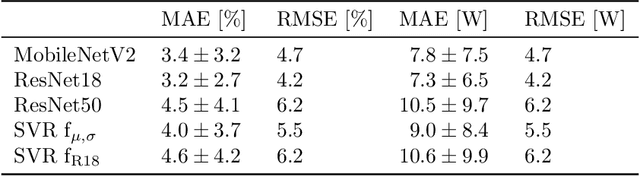
Abstract:Automated inspection plays an important role in monitoring large-scale photovoltaic power plants. Commonly, electroluminescense measurements are used to identify various types of defects on solar modules but have not been used to determine the power of a module. However, knowledge of the power at maximum power point is important as well, since drops in the power of a single module can affect the performance of an entire string. By now, this is commonly determined by measurements that require to discontact or even dismount the module, rendering a regular inspection of individual modules infeasible. In this work, we bridge the gap between electroluminescense measurements and the power determination of a module. We compile a large dataset of 719 electroluminescense measurementsof modules at various stages of degradation, especially cell cracks and fractures, and the corresponding power at maximum power point. Here,we focus on inactive regions and cracks as the predominant type of defect. We set up a baseline regression model to predict the power from electroluminescense measurements with a mean absolute error of 9.0+/-3.7W (4.0+/-8.4%). Then, we show that deep-learning can be used to train a model that performs significantly better (7.3+/-2.7W or 3.2+/-6.5%). With this work, we aim to open a new research topic. Therefore, we publicly release the dataset, the code and trained models to empower other researchers to compare against our results. Finally, we present a thorough evaluation of certain boundary conditions like the dataset size and an automated preprocessing pipeline for on-site measurements showing multiple modules at once.
Projection-to-Projection Translation for Hybrid X-ray and Magnetic Resonance Imaging
Nov 19, 2019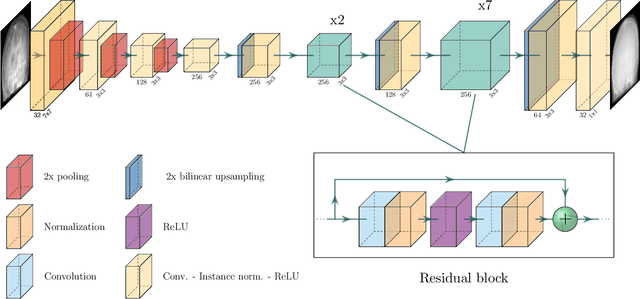
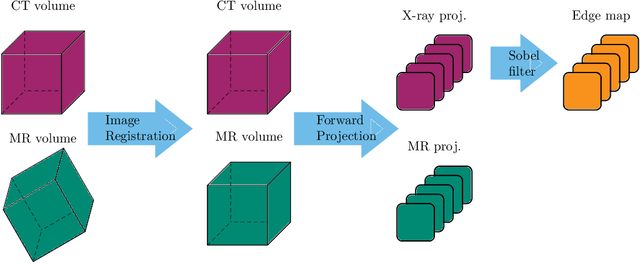
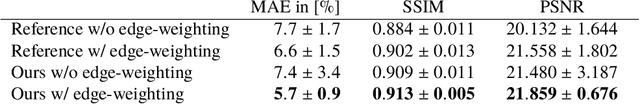
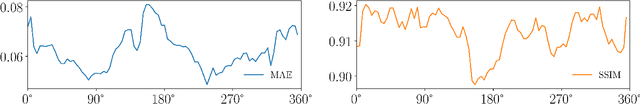
Abstract:Hybrid X-ray and magnetic resonance (MR) imaging promises large potential in interventional medical imaging applications due to the broad variety of contrast of MRI combined with fast imaging of X-ray-based modalities. To fully utilize the potential of the vast amount of existing image enhancement techniques, the corresponding information from both modalities must be present in the same domain. For image-guided interventional procedures, X-ray fluoroscopy has proven to be the modality of choice. Synthesizing one modality from another in this case is an ill-posed problem due to ambiguous signal and overlapping structures in projective geometry. To take on these challenges, we present a learning-based solution to MR to X-ray projection-to-projection translation. We propose an image generator network that focuses on high representation capacity in higher resolution layers to allow for accurate synthesis of fine details in the projection images. Additionally, a weighting scheme in the loss computation that favors high-frequency structures is proposed to focus on the important details and contours in projection imaging. The proposed extensions prove valuable in generating X-ray projection images with natural appearance. Our approach achieves a deviation from the ground truth of only $6$% and structural similarity measure of $0.913\,\pm\,0.005$. In particular the high frequency weighting assists in generating projection images with sharp appearance and reduces erroneously synthesized fine details.
The TCGA Meta-Dataset Clinical Benchmark
Oct 18, 2019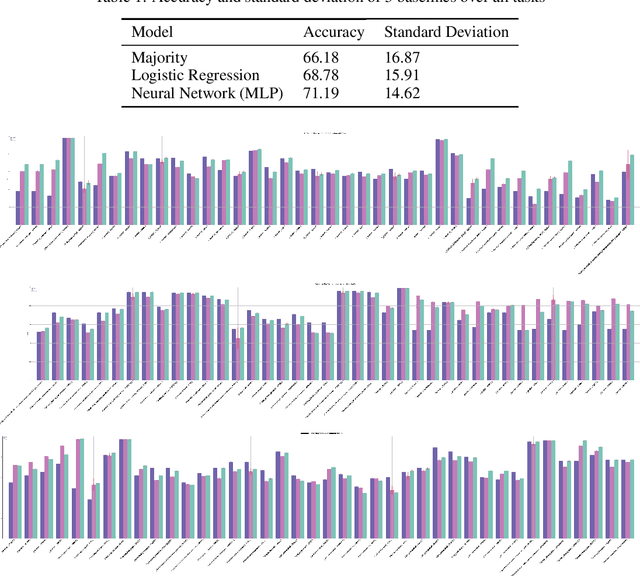
Abstract:Machine learning is bringing a paradigm shift to healthcare by changing the process of disease diagnosis and prognosis in clinics and hospitals. This development equips doctors and medical staff with tools to evaluate their hypotheses and hence make more precise decisions. Although most current research in the literature seeks to develop techniques and methods for predicting one particular clinical outcome, this approach is far from the reality of clinical decision making in which you have to consider several factors simultaneously. In addition, it is difficult to follow the recent progress concretely as there is a lack of consistency in benchmark datasets and task definitions in the field of Genomics. To address the aforementioned issues, we provide a clinical Meta-Dataset derived from the publicly available data hub called The Cancer Genome Atlas Program (TCGA) that contains 174 tasks. We believe those tasks could be good proxy tasks to develop methods which can work on a few samples of gene expression data. Also, learning to predict multiple clinical variables using gene-expression data is an important task due to the variety of phenotypes in clinical problems and lack of samples for some of the rare variables. The defined tasks cover a wide range of clinical problems including predicting tumor tissue site, white cell count, histological type, family history of cancer, gender, and many others which we explain later in the paper. Each task represents an independent dataset. We use regression and neural network baselines for all the tasks using only 150 samples and compare their performance.
Torchmeta: A Meta-Learning library for PyTorch
Sep 14, 2019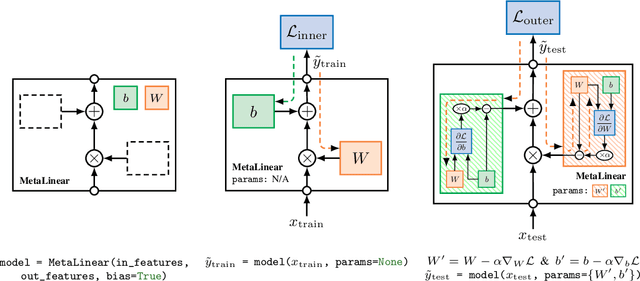
Abstract:The constant introduction of standardized benchmarks in the literature has helped accelerating the recent advances in meta-learning research. They offer a way to get a fair comparison between different algorithms, and the wide range of datasets available allows full control over the complexity of this evaluation. However, for a large majority of code available online, the data pipeline is often specific to one dataset, and testing on another dataset requires significant rework. We introduce Torchmeta, a library built on top of PyTorch that enables seamless and consistent evaluation of meta-learning algorithms on multiple datasets, by providing data-loaders for most of the standard benchmarks in few-shot classification and regression, with a new meta-dataset abstraction. It also features some extensions for PyTorch to simplify the development of models compatible with meta-learning algorithms. The code is available here: https://github.com/tristandeleu/pytorch-meta
Learning with Known Operators reduces Maximum Training Error Bounds
Jul 03, 2019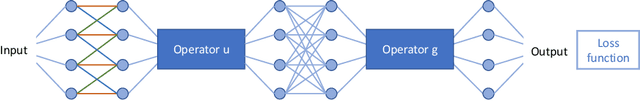
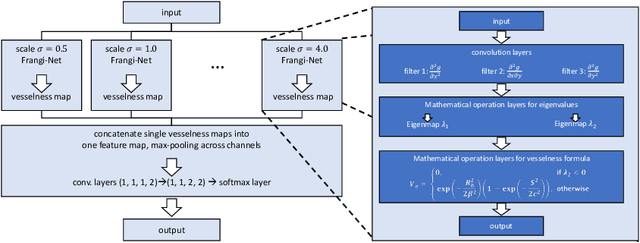
Abstract:We describe an approach for incorporating prior knowledge into machine learning algorithms. We aim at applications in physics and signal processing in which we know that certain operations must be embedded into the algorithm. Any operation that allows computation of a gradient or sub-gradient towards its inputs is suited for our framework. We derive a maximal error bound for deep nets that demonstrates that inclusion of prior knowledge results in its reduction. Furthermore, we also show experimentally that known operators reduce the number of free parameters. We apply this approach to various tasks ranging from CT image reconstruction over vessel segmentation to the derivation of previously unknown imaging algorithms. As such the concept is widely applicable for many researchers in physics, imaging, and signal processing. We assume that our analysis will support further investigation of known operators in other fields of physics, imaging, and signal processing.
Deriving Neural Network Architectures using Precision Learning: Parallel-to-fan beam Conversion
Oct 23, 2018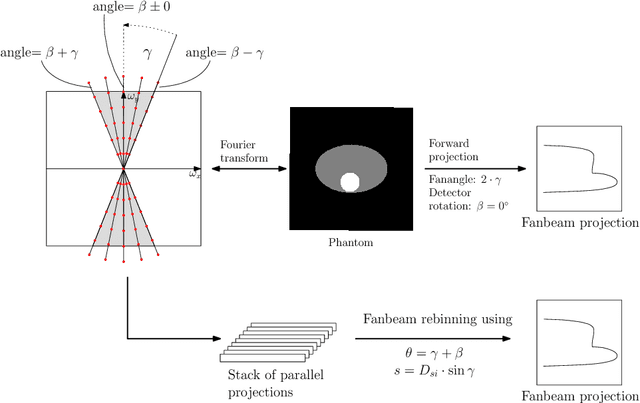
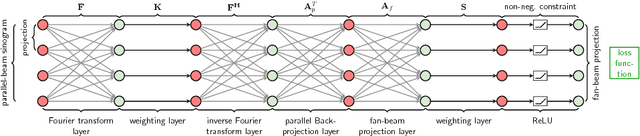
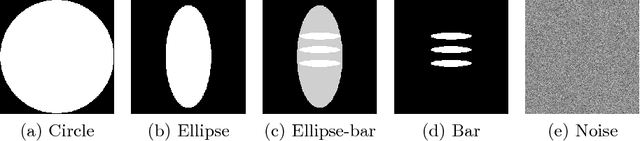
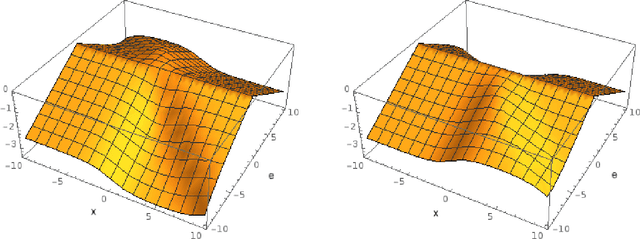
Abstract:In this paper, we derive a neural network architecture based on an analytical formulation of the parallel-to-fan beam conversion problem following the concept of precision learning. The network allows to learn the unknown operators in this conversion in a data-driven manner avoiding interpolation and potential loss of resolution. Integration of known operators results in a small number of trainable parameters that can be estimated from synthetic data only. The concept is evaluated in the context of Hybrid MRI/X-ray imaging where transformation of the parallel-beam MRI projections to fan-beam X-ray projections is required. The proposed method is compared to a traditional rebinning method. The results demonstrate that the proposed method is superior to ray-by-ray interpolation and is able to deliver sharper images using the same amount of parallel-beam input projections which is crucial for interventional applications. We believe that this approach forms a basis for further work uniting deep learning, signal processing, physics, and traditional pattern recognition.
Precision Learning: Towards Use of Known Operators in Neural Networks
Oct 12, 2018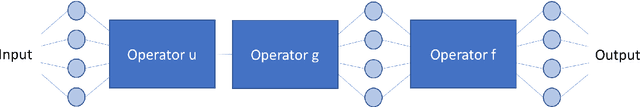
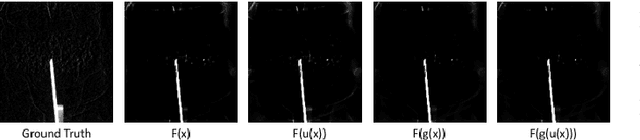
Abstract:In this paper, we consider the use of prior knowledge within neural networks. In particular, we investigate the effect of a known transform within the mapping from input data space to the output domain. We demonstrate that use of known transforms is able to change maximal error bounds. In order to explore the effect further, we consider the problem of X-ray material decomposition as an example to incorporate additional prior knowledge. We demonstrate that inclusion of a non-linear function known from the physical properties of the system is able to reduce prediction errors therewith improving prediction quality from SSIM values of 0.54 to 0.88. This approach is applicable to a wide set of applications in physics and signal processing that provide prior knowledge on such transforms. Also maximal error estimation and network understanding could be facilitated within the context of precision learning.
* accepted on ICPR 2018
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge