Thomas Fevens
Source-free Domain Adaptation Requires Penalized Diversity
Apr 12, 2023



Abstract:While neural networks are capable of achieving human-like performance in many tasks such as image classification, the impressive performance of each model is limited to its own dataset. Source-free domain adaptation (SFDA) was introduced to address knowledge transfer between different domains in the absence of source data, thus, increasing data privacy. Diversity in representation space can be vital to a model`s adaptability in varied and difficult domains. In unsupervised SFDA, the diversity is limited to learning a single hypothesis on the source or learning multiple hypotheses with a shared feature extractor. Motivated by the improved predictive performance of ensembles, we propose a novel unsupervised SFDA algorithm that promotes representational diversity through the use of separate feature extractors with Distinct Backbone Architectures (DBA). Although diversity in feature space is increased, the unconstrained mutual information (MI) maximization may potentially introduce amplification of weak hypotheses. Thus we introduce the Weak Hypothesis Penalization (WHP) regularizer as a mitigation strategy. Our work proposes Penalized Diversity (PD) where the synergy of DBA and WHP is applied to unsupervised source-free domain adaptation for covariate shift. In addition, PD is augmented with a weighted MI maximization objective for label distribution shift. Empirical results on natural, synthetic, and medical domains demonstrate the effectiveness of PD under different distributional shifts.
on the effectiveness of generative adversarial network on anomaly detection
Dec 31, 2021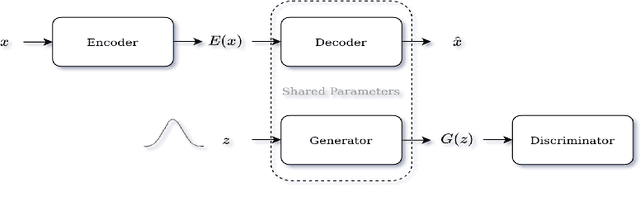
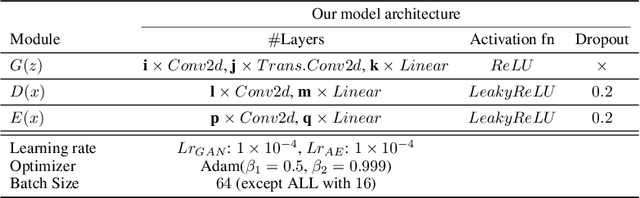
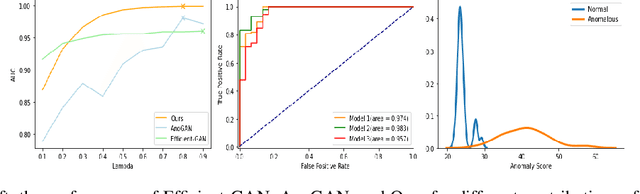
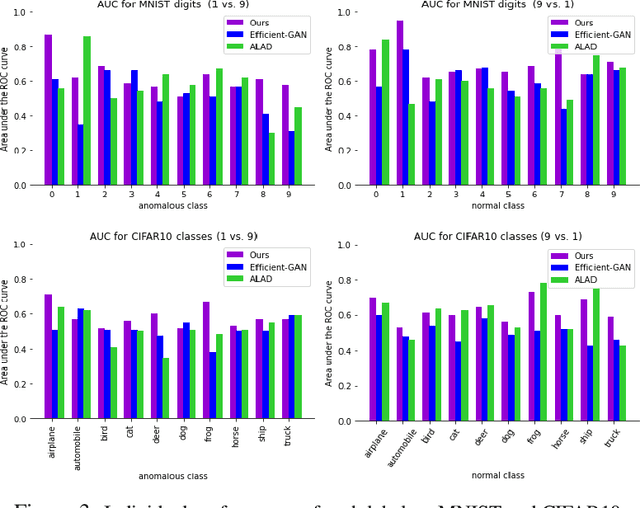
Abstract:Identifying anomalies refers to detecting samples that do not resemble the training data distribution. Many generative models have been used to find anomalies, and among them, generative adversarial network (GAN)-based approaches are currently very popular. GANs mainly rely on the rich contextual information of these models to identify the actual training distribution. Following this analogy, we suggested a new unsupervised model based on GANs --a combination of an autoencoder and a GAN. Further, a new scoring function was introduced to target anomalies where a linear combination of the internal representation of the discriminator and the generator's visual representation, plus the encoded representation of the autoencoder, come together to define the proposed anomaly score. The model was further evaluated on benchmark datasets such as SVHN, CIFAR10, and MNIST, as well as a public medical dataset of leukemia images. In all the experiments, our model outperformed its existing counterparts while slightly improving the inference time.
Boosting Segmentation Performance across datasets using histogram specification with application to pelvic bone segmentation
Jan 26, 2021



Abstract:Accurate segmentation of the pelvic CTs is crucial for the clinical diagnosis of pelvic bone diseases and for planning patient-specific hip surgeries. With the emergence and advancements of deep learning for digital healthcare, several methodologies have been proposed for such segmentation tasks. But in a low data scenario, the lack of abundant data needed to train a Deep Neural Network is a significant bottle-neck. In this work, we propose a methodology based on modulation of image tonal distributions and deep learning to boost the performance of networks trained on limited data. The strategy involves pre-processing of test data through histogram specification. This simple yet effective approach can be viewed as a style transfer methodology. The segmentation task uses a U-Net configuration with an EfficientNet-B0 backbone, optimized using an augmented BCE-IoU loss function. This configuration is validated on a total of 284 images taken from two publicly available CT datasets, TCIA (a cancer imaging archive) and the Visible Human Project. The average performance measures for the Dice coefficient and Intersection over Union are 95.7% and 91.9%, respectively, give strong evidence for the effectiveness of the approach, which is highly competitive with state-of-the-art methodologies.
FoCL: Feature-Oriented Continual Learning for Generative Models
Mar 09, 2020



Abstract:In this paper, we propose a general framework in continual learning for generative models: Feature-oriented Continual Learning (FoCL). Unlike previous works that aim to solve the catastrophic forgetting problem by introducing regularization in the parameter space or image space, FoCL imposes regularization in the feature space. We show in our experiments that FoCL has faster adaptation to distributional changes in sequentially arriving tasks, and achieves the state-of-the-art performance for generative models in task incremental learning. We discuss choices of combined regularization spaces towards different use case scenarios for boosted performance, e.g., tasks that have high variability in the background. Finally, we introduce a forgetfulness measure that fairly evaluates the degree to which a model suffers from forgetting. Interestingly, the analysis of our proposed forgetfulness score also implies that FoCL tends to have a mitigated forgetting for future tasks.
The TCGA Meta-Dataset Clinical Benchmark
Oct 18, 2019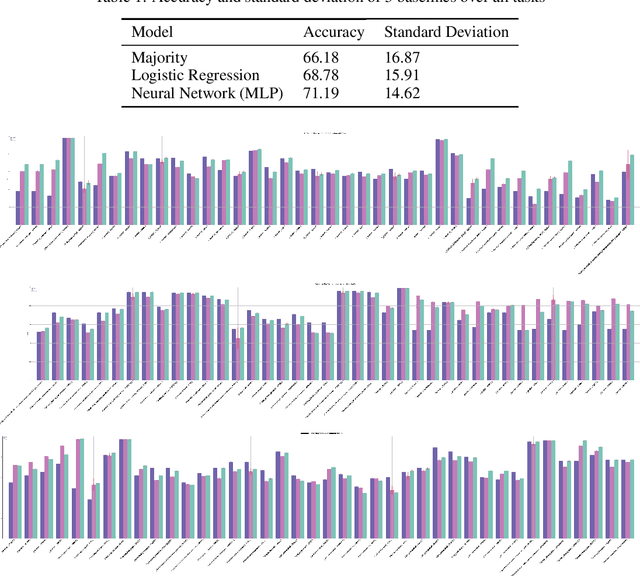
Abstract:Machine learning is bringing a paradigm shift to healthcare by changing the process of disease diagnosis and prognosis in clinics and hospitals. This development equips doctors and medical staff with tools to evaluate their hypotheses and hence make more precise decisions. Although most current research in the literature seeks to develop techniques and methods for predicting one particular clinical outcome, this approach is far from the reality of clinical decision making in which you have to consider several factors simultaneously. In addition, it is difficult to follow the recent progress concretely as there is a lack of consistency in benchmark datasets and task definitions in the field of Genomics. To address the aforementioned issues, we provide a clinical Meta-Dataset derived from the publicly available data hub called The Cancer Genome Atlas Program (TCGA) that contains 174 tasks. We believe those tasks could be good proxy tasks to develop methods which can work on a few samples of gene expression data. Also, learning to predict multiple clinical variables using gene-expression data is an important task due to the variety of phenotypes in clinical problems and lack of samples for some of the rare variables. The defined tasks cover a wide range of clinical problems including predicting tumor tissue site, white cell count, histological type, family history of cancer, gender, and many others which we explain later in the paper. Each task represents an independent dataset. We use regression and neural network baselines for all the tasks using only 150 samples and compare their performance.
Dual Adversarial Inference for Text-to-Image Synthesis
Aug 14, 2019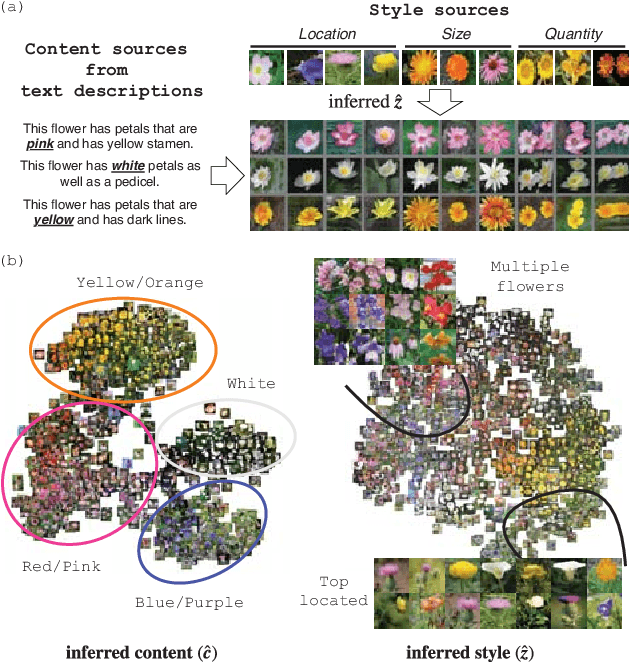

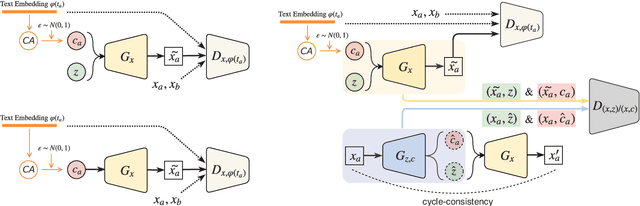
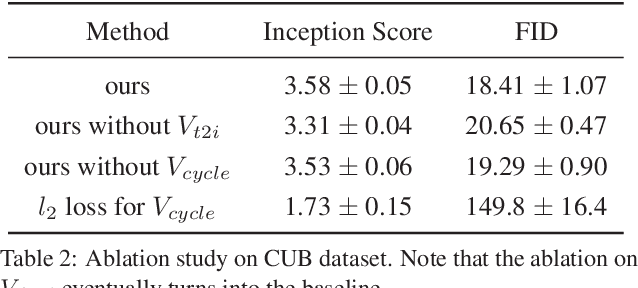
Abstract:Synthesizing images from a given text description involves engaging two types of information: the content, which includes information explicitly described in the text (e.g., color, composition, etc.), and the style, which is usually not well described in the text (e.g., location, quantity, size, etc.). However, in previous works, it is typically treated as a process of generating images only from the content, i.e., without considering learning meaningful style representations. In this paper, we aim to learn two variables that are disentangled in the latent space, representing content and style respectively. We achieve this by augmenting current text-to-image synthesis frameworks with a dual adversarial inference mechanism. Through extensive experiments, we show that our model learns, in an unsupervised manner, style representations corresponding to certain meaningful information present in the image that are not well described in the text. The new framework also improves the quality of synthesized images when evaluated on Oxford-102, CUB and COCO datasets.
Case-Based Histopathological Malignancy Diagnosis using Convolutional Neural Networks
May 28, 2019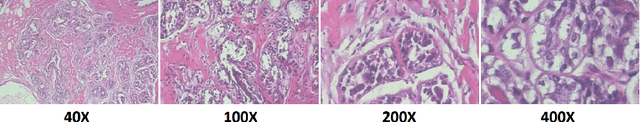

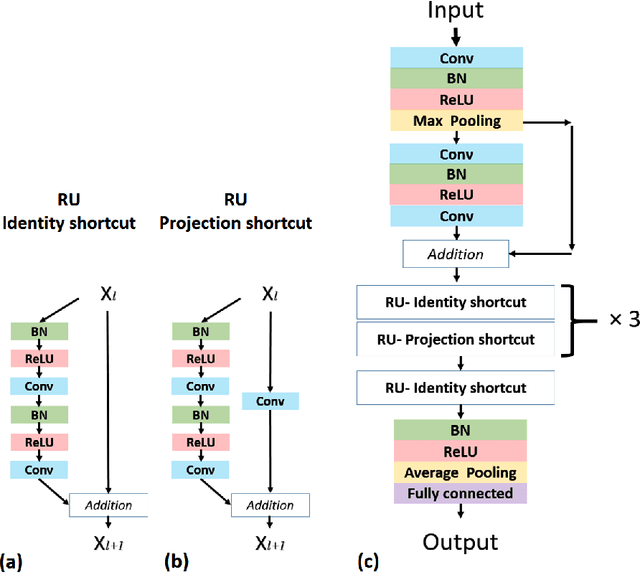
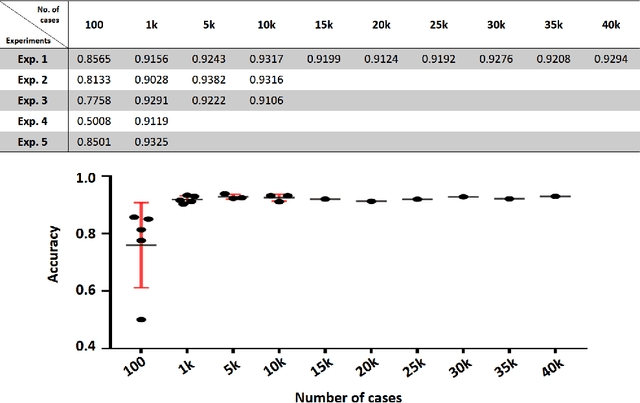
Abstract:In practice, histopathological diagnosis of tumor malignancy often requires a human expert to scan through histopathological images at multiple magnification levels, after which a final diagnosis can be accurately determined. However, previous research on such classification tasks using convolutional neural networks primarily determine a diagnosis for a single magnification level. In this paper, we propose a case-based approach using deep residual neural networks for histopathological malignancy diagnosis, where a case is defined as a sequence of images from the patient at all available levels of magnification. Effectively, through mimicking what a human expert would actually do, our approach makes a diagnosis decision based on features learned in combination at multiple magnification levels. Our results show that the case-based approach achieves better performance than the state-of-the-art methods when evaluated on BreaKHis, a histopathological image dataset for breast tumors.
Leveraging Disease Progression Learning for Medical Image Recognition
Sep 01, 2018



Abstract:Unlike natural images, medical images often have intrinsic characteristics that can be leveraged for neural network learning. For example, images that belong to different stages of a disease may continuously follow a certain progression pattern. In this paper, we propose a novel method that leverages disease progression learning for medical image recognition. In our method, sequences of images ordered by disease stages are learned by a neural network that consists of a shared vision model for feature extraction and a long short-term memory network for the learning of stage sequences. Auxiliary vision outputs are also included to capture stage features that tend to be discrete along the disease progression. Our proposed method is evaluated on a public diabetic retinopathy dataset, and achieves about 3.3% improvement in disease staging accuracy, compared to the baseline method that does not use disease progression learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge