Francis Dutil
Application of Homomorphic Encryption in Medical Imaging
Oct 12, 2021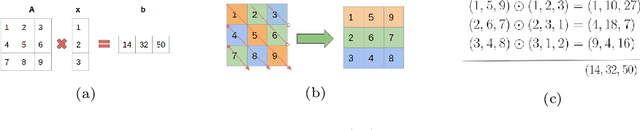
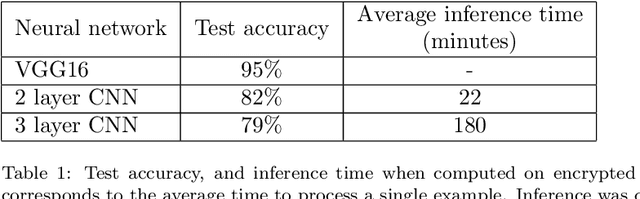
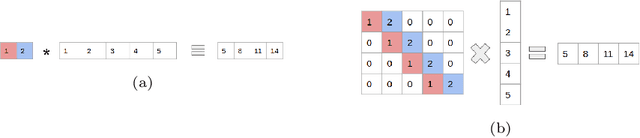
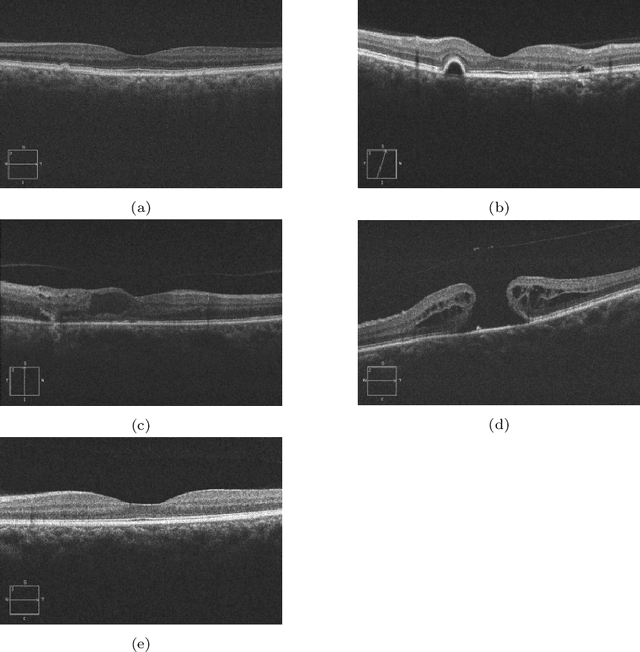
Abstract:In this technical report, we explore the use of homomorphic encryption (HE) in the context of training and predicting with deep learning (DL) models to deliver strict \textit{Privacy by Design} services, and to enforce a zero-trust model of data governance. First, we show how HE can be used to make predictions over medical images while preventing unauthorized secondary use of data, and detail our results on a disease classification task with OCT images. Then, we demonstrate that HE can be used to secure the training of DL models through federated learning, and report some experiments using 3D chest CT-Scans for a nodule detection task.
Cross-Modal Information Maximization for Medical Imaging: CMIM
Oct 20, 2020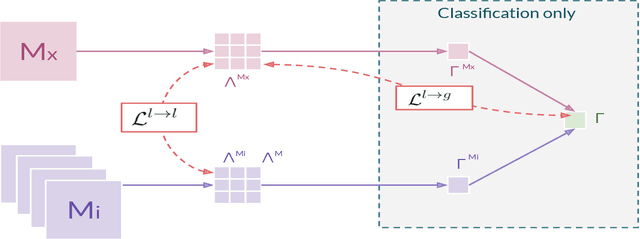

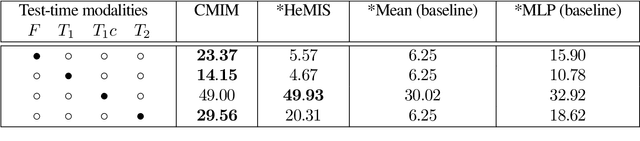
Abstract:In hospitals, data are siloed to specific information systems that make the same information available under different modalities such as the different medical imaging exams the patient undergoes (CT scans, MRI, PET, Ultrasound, etc.) and their associated radiology reports. This offers unique opportunities to obtain and use at train-time those multiple views of the same information that might not always be available at test-time. In this paper, we propose an innovative framework that makes the most of available data by learning good representations of a multi-modal input that are resilient to modality dropping at test-time, using recent advances in mutual information maximization. By maximizing cross-modal information at train time, we are able to outperform several state-of-the-art baselines in two different settings, medical image classification, and segmentation. In particular, our method is shown to have a strong impact on the inference-time performance of weaker modalities.
FoCL: Feature-Oriented Continual Learning for Generative Models
Mar 09, 2020



Abstract:In this paper, we propose a general framework in continual learning for generative models: Feature-oriented Continual Learning (FoCL). Unlike previous works that aim to solve the catastrophic forgetting problem by introducing regularization in the parameter space or image space, FoCL imposes regularization in the feature space. We show in our experiments that FoCL has faster adaptation to distributional changes in sequentially arriving tasks, and achieves the state-of-the-art performance for generative models in task incremental learning. We discuss choices of combined regularization spaces towards different use case scenarios for boosted performance, e.g., tasks that have high variability in the background. Finally, we introduce a forgetfulness measure that fairly evaluates the degree to which a model suffers from forgetting. Interestingly, the analysis of our proposed forgetfulness score also implies that FoCL tends to have a mitigated forgetting for future tasks.
The TCGA Meta-Dataset Clinical Benchmark
Oct 18, 2019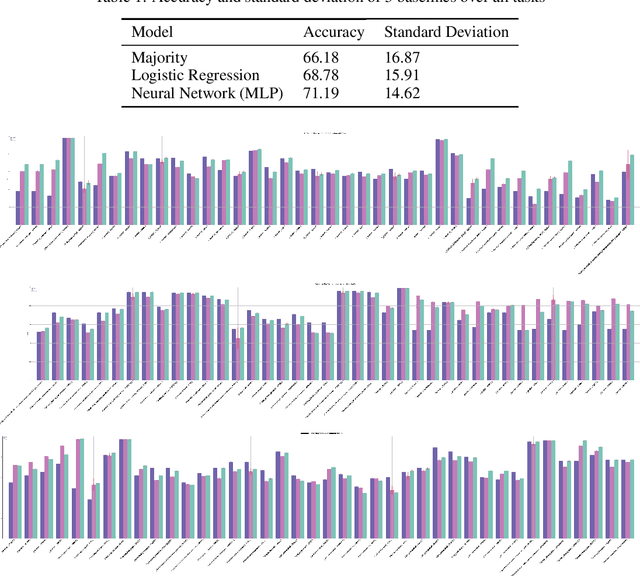
Abstract:Machine learning is bringing a paradigm shift to healthcare by changing the process of disease diagnosis and prognosis in clinics and hospitals. This development equips doctors and medical staff with tools to evaluate their hypotheses and hence make more precise decisions. Although most current research in the literature seeks to develop techniques and methods for predicting one particular clinical outcome, this approach is far from the reality of clinical decision making in which you have to consider several factors simultaneously. In addition, it is difficult to follow the recent progress concretely as there is a lack of consistency in benchmark datasets and task definitions in the field of Genomics. To address the aforementioned issues, we provide a clinical Meta-Dataset derived from the publicly available data hub called The Cancer Genome Atlas Program (TCGA) that contains 174 tasks. We believe those tasks could be good proxy tasks to develop methods which can work on a few samples of gene expression data. Also, learning to predict multiple clinical variables using gene-expression data is an important task due to the variety of phenotypes in clinical problems and lack of samples for some of the rare variables. The defined tasks cover a wide range of clinical problems including predicting tumor tissue site, white cell count, histological type, family history of cancer, gender, and many others which we explain later in the paper. Each task represents an independent dataset. We use regression and neural network baselines for all the tasks using only 150 samples and compare their performance.
Underwhelming Generalization Improvements From Controlling Feature Attribution
Oct 01, 2019



Abstract:Overfitting is a common issue in machine learning, which can arise when the model learns to predict class membership using convenient but spuriously-correlated image features instead of the true image features that denote a class. These are typically visualized using saliency maps. In some object classification tasks such as for medical images, one may have some images with masks, indicating a region of interest, i.e., which part of the image contains the most relevant information for the classification. We describe a simple method for taking advantage of such auxiliary labels, by training networks to ignore the distracting features which may be extracted outside of the region of interest, on the training images for which such masks are available. This mask information is only used during training and has an impact on generalization accuracy in a dataset-dependent way. We observe an underwhelming relationship between controlling saliency maps and improving generalization performance.
Dual Adversarial Inference for Text-to-Image Synthesis
Aug 14, 2019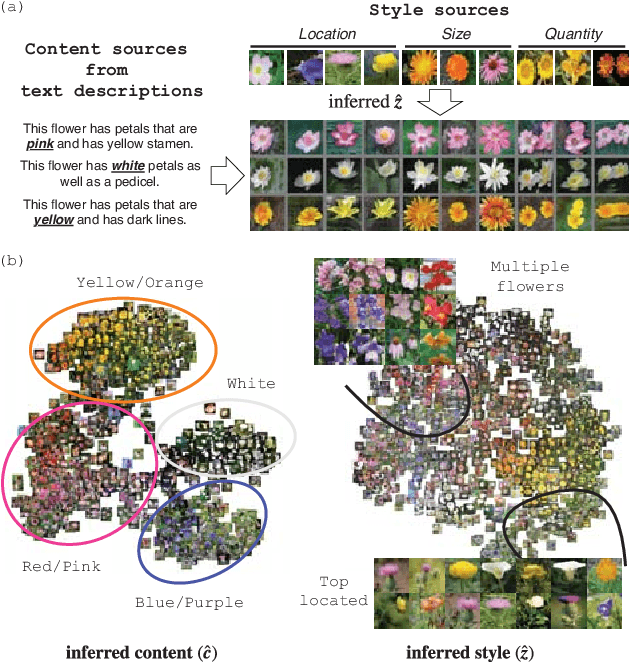

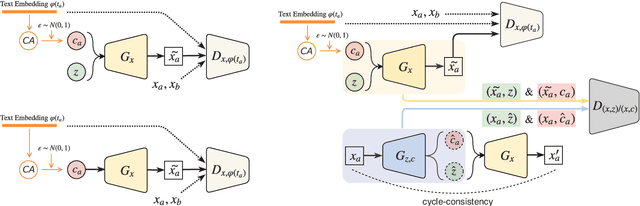
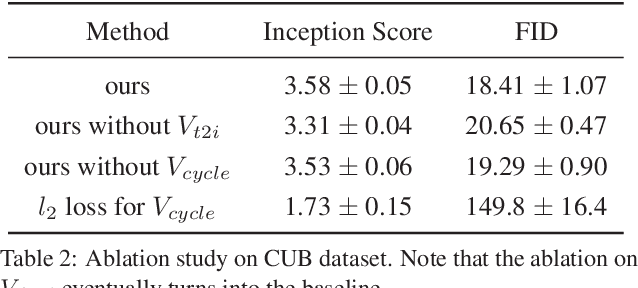
Abstract:Synthesizing images from a given text description involves engaging two types of information: the content, which includes information explicitly described in the text (e.g., color, composition, etc.), and the style, which is usually not well described in the text (e.g., location, quantity, size, etc.). However, in previous works, it is typically treated as a process of generating images only from the content, i.e., without considering learning meaningful style representations. In this paper, we aim to learn two variables that are disentangled in the latent space, representing content and style respectively. We achieve this by augmenting current text-to-image synthesis frameworks with a dual adversarial inference mechanism. Through extensive experiments, we show that our model learns, in an unsupervised manner, style representations corresponding to certain meaningful information present in the image that are not well described in the text. The new framework also improves the quality of synthesized images when evaluated on Oxford-102, CUB and COCO datasets.
GradMask: Reduce Overfitting by Regularizing Saliency
Apr 16, 2019

Abstract:With too few samples or too many model parameters, overfitting can inhibit the ability to generalise predictions to new data. Within medical imaging, this can occur when features are incorrectly assigned importance such as distinct hospital specific artifacts, leading to poor performance on a new dataset from a different institution without those features, which is undesirable. Most regularization methods do not explicitly penalize the incorrect association of these features to the target class and hence fail to address this issue. We propose a regularization method, GradMask, which penalizes saliency maps inferred from the classifier gradients when they are not consistent with the lesion segmentation. This prevents non-tumor related features to contribute to the classification of unhealthy samples. We demonstrate that this method can improve test accuracy between 1-3% compared to the baseline without GradMask, showing that it has an impact on reducing overfitting.
InfoMask: Masked Variational Latent Representation to Localize Chest Disease
Mar 28, 2019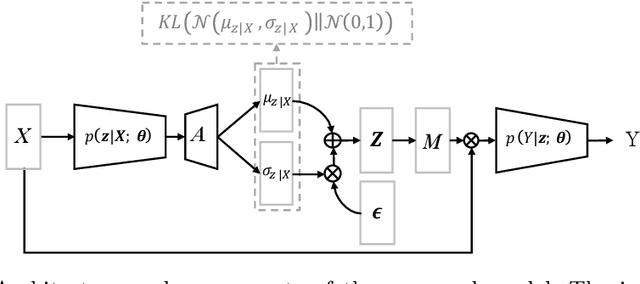
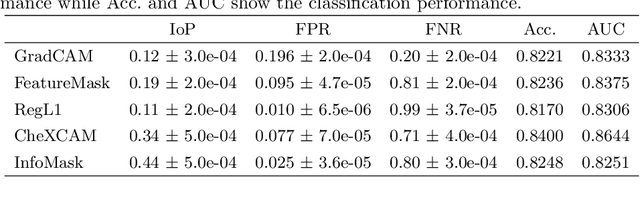

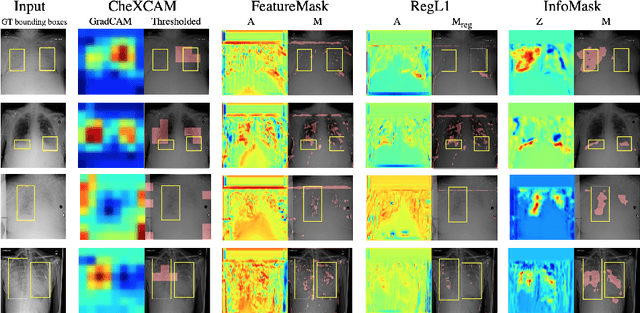
Abstract:The scarcity of richly annotated medical images is limiting supervised deep learning based solutions to medical image analysis tasks, such as localizing discriminatory radiomic disease signatures. Therefore, it is desirable to leverage unsupervised and weakly supervised models. Most recent weakly supervised localization methods apply attention maps or region proposals in a multiple instance learning formulation. While attention maps can be noisy, leading to erroneously highlighted regions, it is not simple to decide on an optimal window/bag size for multiple instance learning approaches. In this paper, we propose a learned spatial masking mechanism to filter out irrelevant background signals from attention maps. The proposed method minimizes mutual information between a masked variational representation and the input while maximizing the information between the masked representation and class labels. This results in more accurate localization of discriminatory regions. We tested the proposed model on the ChestX-ray8 dataset to localize pneumonia from chest X-ray images without using any pixel-level or bounding-box annotations.
Towards the Latent Transcriptome
Oct 08, 2018



Abstract:In this work we propose a method to compute continuous embeddings for kmers from raw RNA-seq data, in a reference-free fashion. We report that our model captures information of both DNA sequence similarity as well as DNA sequence abundance in the embedding latent space. We confirm the quality of these vectors by comparing them to known gene sub-structures and report that the latent space recovers exon information from raw RNA-Seq data from acute myeloid leukemia patients. Furthermore we show that this latent space allows the detection of genomic abnormalities such as translocations as well as patient-specific mutations, making this representation space both useful for visualization as well as analysis.
Towards Gene Expression Convolutions using Gene Interaction Graphs
Jun 18, 2018



Abstract:We study the challenges of applying deep learning to gene expression data. We find experimentally that there exists non-linear signal in the data, however is it not discovered automatically given the noise and low numbers of samples used in most research. We discuss how gene interaction graphs (same pathway, protein-protein, co-expression, or research paper text association) can be used to impose a bias on a deep model similar to the spatial bias imposed by convolutions on an image. We explore the usage of Graph Convolutional Neural Networks coupled with dropout and gene embeddings to utilize the graph information. We find this approach provides an advantage for particular tasks in a low data regime but is very dependent on the quality of the graph used. We conclude that more work should be done in this direction. We design experiments that show why existing methods fail to capture signal that is present in the data when features are added which clearly isolates the problem that needs to be addressed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge