Claude Perreault
Institute for Research in Immunology and Cancer, Department of Medicine, Université de Montréal
Towards the Latent Transcriptome
Oct 08, 2018



Abstract:In this work we propose a method to compute continuous embeddings for kmers from raw RNA-seq data, in a reference-free fashion. We report that our model captures information of both DNA sequence similarity as well as DNA sequence abundance in the embedding latent space. We confirm the quality of these vectors by comparing them to known gene sub-structures and report that the latent space recovers exon information from raw RNA-Seq data from acute myeloid leukemia patients. Furthermore we show that this latent space allows the detection of genomic abnormalities such as translocations as well as patient-specific mutations, making this representation space both useful for visualization as well as analysis.
Holographic Neural Architectures
Jun 04, 2018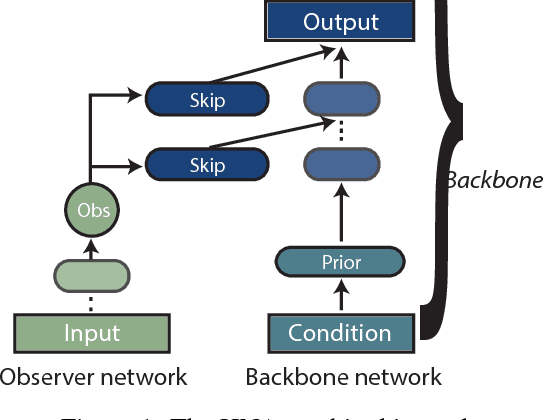

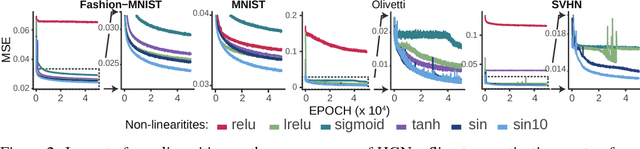

Abstract:Representation learning is at the heart of what makes deep learning effective. In this work, we introduce a new framework for representation learning that we call "Holographic Neural Architectures" (HNAs). In the same way that an observer can experience the 3D structure of a holographed object by looking at its hologram from several angles, HNAs derive Holographic Representations from the training set. These representations can then be explored by moving along a continuous bounded single dimension. We show that HNAs can be used to make generative networks, state-of-the-art regression models and that they are inherently highly resistant to noise. Finally, we argue that because of their denoising abilities and their capacity to generalize well from very few examples, models based upon HNAs are particularly well suited for biological applications where training examples are rare or noisy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge