Assya Trofimov
Accounting for Variance in Machine Learning Benchmarks
Mar 01, 2021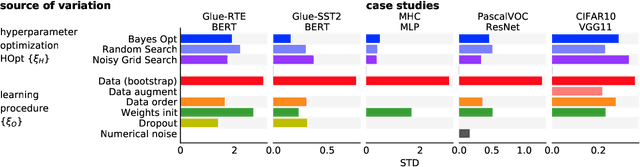
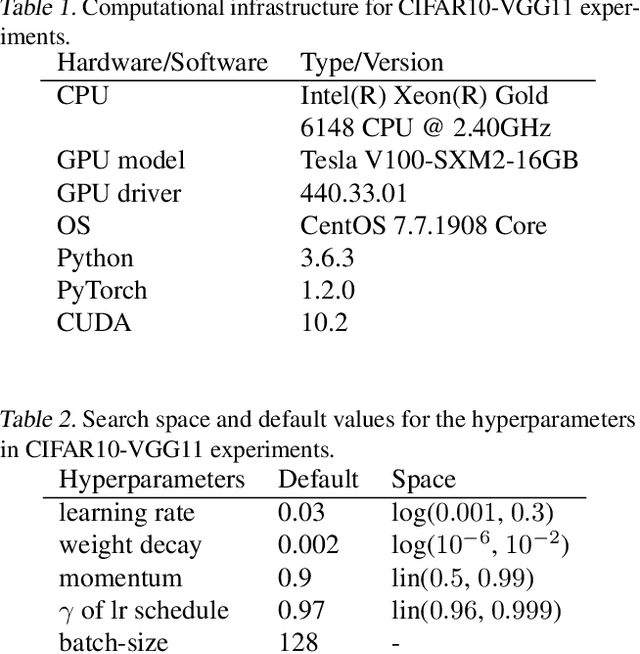
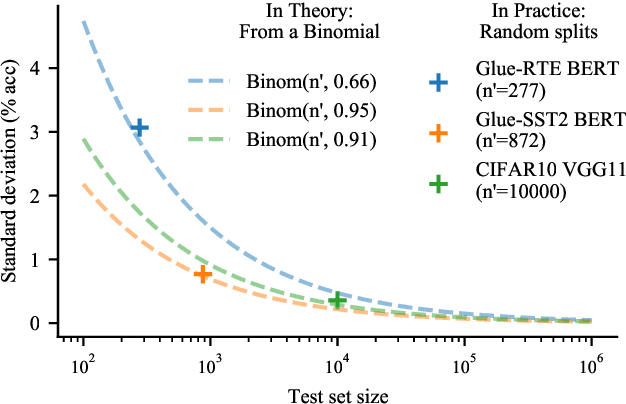
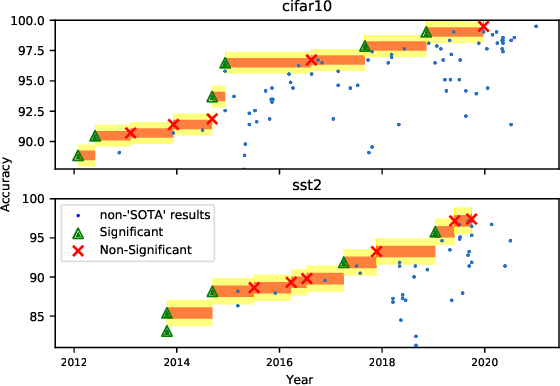
Abstract:Strong empirical evidence that one machine-learning algorithm A outperforms another one B ideally calls for multiple trials optimizing the learning pipeline over sources of variation such as data sampling, data augmentation, parameter initialization, and hyperparameters choices. This is prohibitively expensive, and corners are cut to reach conclusions. We model the whole benchmarking process, revealing that variance due to data sampling, parameter initialization and hyperparameter choice impact markedly the results. We analyze the predominant comparison methods used today in the light of this variance. We show a counter-intuitive result that adding more sources of variation to an imperfect estimator approaches better the ideal estimator at a 51 times reduction in compute cost. Building on these results, we study the error rate of detecting improvements, on five different deep-learning tasks/architectures. This study leads us to propose recommendations for performance comparisons.
Towards the Latent Transcriptome
Oct 08, 2018



Abstract:In this work we propose a method to compute continuous embeddings for kmers from raw RNA-seq data, in a reference-free fashion. We report that our model captures information of both DNA sequence similarity as well as DNA sequence abundance in the embedding latent space. We confirm the quality of these vectors by comparing them to known gene sub-structures and report that the latent space recovers exon information from raw RNA-Seq data from acute myeloid leukemia patients. Furthermore we show that this latent space allows the detection of genomic abnormalities such as translocations as well as patient-specific mutations, making this representation space both useful for visualization as well as analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge