Justin Szeto
KerJEPA: Kernel Discrepancies for Euclidean Self-Supervised Learning
Dec 22, 2025Abstract:Recent breakthroughs in self-supervised Joint-Embedding Predictive Architectures (JEPAs) have established that regularizing Euclidean representations toward isotropic Gaussian priors yields provable gains in training stability and downstream generalization. We introduce a new, flexible family of KerJEPAs, self-supervised learning algorithms with kernel-based regularizers. One instance of this family corresponds to the recently-introduced LeJEPA Epps-Pulley regularizer which approximates a sliced maximum mean discrepancy (MMD) with a Gaussian prior and Gaussian kernel. By expanding the class of viable kernels and priors and computing the closed-form high-dimensional limit of sliced MMDs, we develop alternative KerJEPAs with a number of favorable properties including improved training stability and design flexibility.
Spatio-Temporal Conditional Diffusion Models for Forecasting Future Multiple Sclerosis Lesion Masks Conditioned on Treatments
Aug 09, 2025Abstract:Image-based personalized medicine has the potential to transform healthcare, particularly for diseases that exhibit heterogeneous progression such as Multiple Sclerosis (MS). In this work, we introduce the first treatment-aware spatio-temporal diffusion model that is able to generate future masks demonstrating lesion evolution in MS. Our voxel-space approach incorporates multi-modal patient data, including MRI and treatment information, to forecast new and enlarging T2 (NET2) lesion masks at a future time point. Extensive experiments on a multi-centre dataset of 2131 patient 3D MRIs from randomized clinical trials for relapsing-remitting MS demonstrate that our generative model is able to accurately predict NET2 lesion masks for patients across six different treatments. Moreover, we demonstrate our model has the potential for real-world clinical applications through downstream tasks such as future lesion count and location estimation, binary lesion activity classification, and generating counterfactual future NET2 masks for several treatments with different efficacies. This work highlights the potential of causal, image-based generative models as powerful tools for advancing data-driven prognostics in MS.
Benchmarking a Benchmark: How Reliable is MS-COCO?
Nov 05, 2023Abstract:Benchmark datasets are used to profile and compare algorithms across a variety of tasks, ranging from image classification to segmentation, and also play a large role in image pretraining algorithms. Emphasis is placed on results with little regard to the actual content within the dataset. It is important to question what kind of information is being learned from these datasets and what are the nuances and biases within them. In the following work, Sama-COCO, a re-annotation of MS-COCO, is used to discover potential biases by leveraging a shape analysis pipeline. A model is trained and evaluated on both datasets to examine the impact of different annotation conditions. Results demonstrate that annotation styles are important and that annotation pipelines should closely consider the task of interest. The dataset is made publicly available at https://www.sama.com/sama-coco-dataset/ .
An Empirical Study of Uncertainty in Polygon Annotation and the Impact of Quality Assurance
Nov 05, 2023



Abstract:Polygons are a common annotation format used for quickly annotating objects in instance segmentation tasks. However, many real-world annotation projects request near pixel-perfect labels. While strict pixel guidelines may appear to be the solution to a successful project, practitioners often fail to assess the feasibility of the work requested, and overlook common factors that may challenge the notion of quality. This paper aims to examine and quantify the inherent uncertainty for polygon annotations and the role that quality assurance plays in minimizing its effect. To this end, we conduct an analysis on multi-rater polygon annotations for several objects from the MS-COCO dataset. The results demonstrate that the reliability of a polygon annotation is dependent on a reviewing procedure, as well as the scene and shape complexity.
Mitigating Calibration Bias Without Fixed Attribute Grouping for Improved Fairness in Medical Imaging Analysis
Jul 20, 2023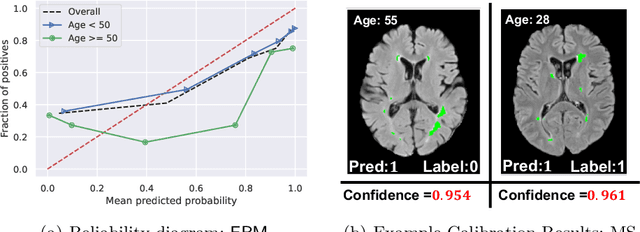
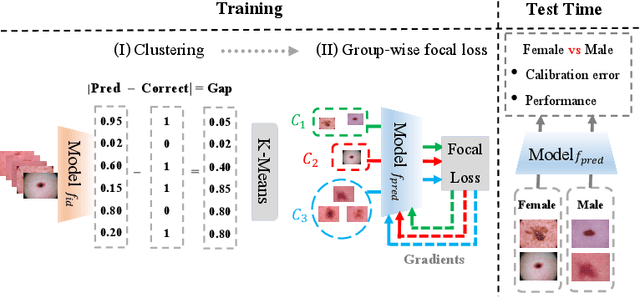

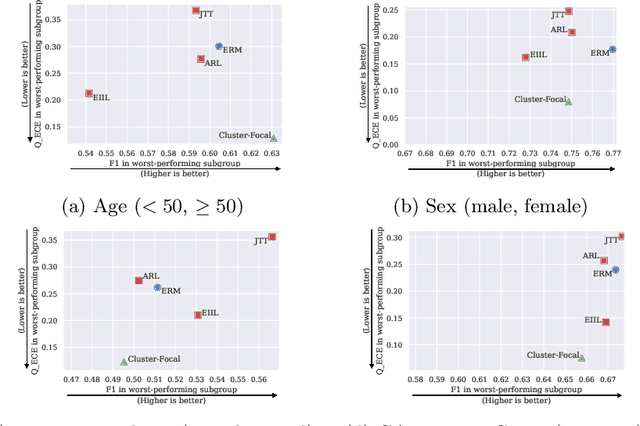
Abstract:Trustworthy deployment of deep learning medical imaging models into real-world clinical practice requires that they be calibrated. However, models that are well calibrated overall can still be poorly calibrated for a sub-population, potentially resulting in a clinician unwittingly making poor decisions for this group based on the recommendations of the model. Although methods have been shown to successfully mitigate biases across subgroups in terms of model accuracy, this work focuses on the open problem of mitigating calibration biases in the context of medical image analysis. Our method does not require subgroup attributes during training, permitting the flexibility to mitigate biases for different choices of sensitive attributes without re-training. To this end, we propose a novel two-stage method: Cluster-Focal to first identify poorly calibrated samples, cluster them into groups, and then introduce group-wise focal loss to improve calibration bias. We evaluate our method on skin lesion classification with the public HAM10000 dataset, and on predicting future lesional activity for multiple sclerosis (MS) patients. In addition to considering traditional sensitive attributes (e.g. age, sex) with demographic subgroups, we also consider biases among groups with different image-derived attributes, such as lesion load, which are required in medical image analysis. Our results demonstrate that our method effectively controls calibration error in the worst-performing subgroups while preserving prediction performance, and outperforming recent baselines.
Rethinking Generalization: The Impact of Annotation Style on Medical Image Segmentation
Oct 31, 2022Abstract:Generalization is an important attribute of machine learning models, particularly for those that are to be deployed in a medical context, where unreliable predictions can have real world consequences. While the failure of models to generalize across datasets is typically attributed to a mismatch in the data distributions, performance gaps are often a consequence of biases in the ``ground-truth" label annotations. This is particularly important in the context of medical image segmentation of pathological structures (e.g. lesions), where the annotation process is much more subjective, and affected by a number underlying factors, including the annotation protocol, rater education/experience, and clinical aims, among others. In this paper, we show that modeling annotation biases, rather than ignoring them, poses a promising way of accounting for differences in annotation style across datasets. To this end, we propose a generalized conditioning framework to (1) learn and account for different annotation styles across multiple datasets using a single model, (2) identify similar annotation styles across different datasets in order to permit their effective aggregation, and (3) fine-tune a fully trained model to a new annotation style with just a few samples. Next, we present an image-conditioning approach to model annotation styles that correlate with specific image features, potentially enabling detection biases to be more easily identified.
Cohort Bias Adaptation in Aggregated Datasets for Lesion Segmentation
Aug 02, 2021



Abstract:Many automatic machine learning models developed for focal pathology (e.g. lesions, tumours) detection and segmentation perform well, but do not generalize as well to new patient cohorts, impeding their widespread adoption into real clinical contexts. One strategy to create a more diverse, generalizable training set is to naively pool datasets from different cohorts. Surprisingly, training on this \it{big data} does not necessarily increase, and may even reduce, overall performance and model generalizability, due to the existence of cohort biases that affect label distributions. In this paper, we propose a generalized affine conditioning framework to learn and account for cohort biases across multi-source datasets, which we call Source-Conditioned Instance Normalization (SCIN). Through extensive experimentation on three different, large scale, multi-scanner, multi-centre Multiple Sclerosis (MS) clinical trial MRI datasets, we show that our cohort bias adaptation method (1) improves performance of the network on pooled datasets relative to naively pooling datasets and (2) can quickly adapt to a new cohort by fine-tuning the instance normalization parameters, thus learning the new cohort bias with only 10 labelled samples.
Optimizing Operating Points for High Performance Lesion Detection and Segmentation Using Lesion Size Reweighting
Jul 27, 2021
Abstract:There are many clinical contexts which require accurate detection and segmentation of all focal pathologies (e.g. lesions, tumours) in patient images. In cases where there are a mix of small and large lesions, standard binary cross entropy loss will result in better segmentation of large lesions at the expense of missing small ones. Adjusting the operating point to accurately detect all lesions generally leads to oversegmentation of large lesions. In this work, we propose a novel reweighing strategy to eliminate this performance gap, increasing small pathology detection performance while maintaining segmentation accuracy. We show that our reweighing strategy vastly outperforms competing strategies based on experiments on a large scale, multi-scanner, multi-center dataset of Multiple Sclerosis patient images.
Accounting for Variance in Machine Learning Benchmarks
Mar 01, 2021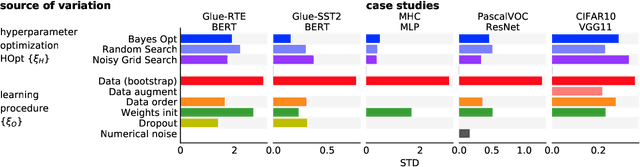
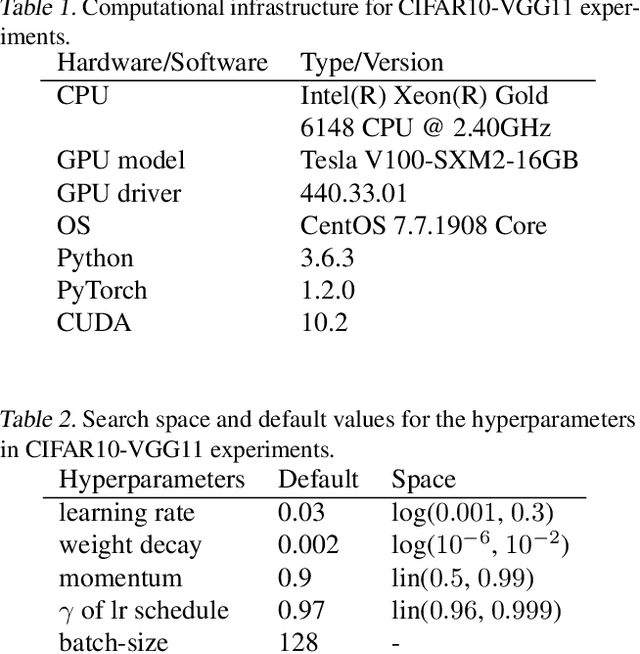
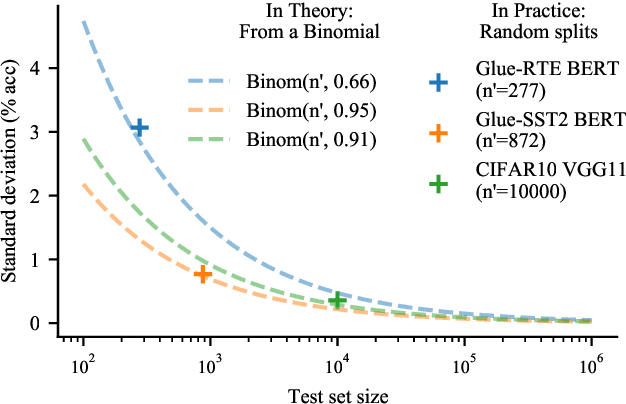
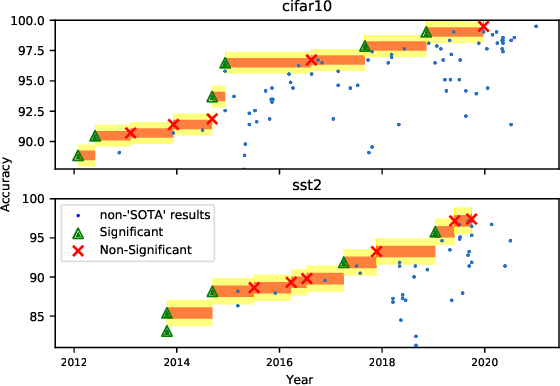
Abstract:Strong empirical evidence that one machine-learning algorithm A outperforms another one B ideally calls for multiple trials optimizing the learning pipeline over sources of variation such as data sampling, data augmentation, parameter initialization, and hyperparameters choices. This is prohibitively expensive, and corners are cut to reach conclusions. We model the whole benchmarking process, revealing that variance due to data sampling, parameter initialization and hyperparameter choice impact markedly the results. We analyze the predominant comparison methods used today in the light of this variance. We show a counter-intuitive result that adding more sources of variation to an imperfect estimator approaches better the ideal estimator at a 51 times reduction in compute cost. Building on these results, we study the error rate of detecting improvements, on five different deep-learning tasks/architectures. This study leads us to propose recommendations for performance comparisons.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge