Eric Zimmermann
KerJEPA: Kernel Discrepancies for Euclidean Self-Supervised Learning
Dec 22, 2025Abstract:Recent breakthroughs in self-supervised Joint-Embedding Predictive Architectures (JEPAs) have established that regularizing Euclidean representations toward isotropic Gaussian priors yields provable gains in training stability and downstream generalization. We introduce a new, flexible family of KerJEPAs, self-supervised learning algorithms with kernel-based regularizers. One instance of this family corresponds to the recently-introduced LeJEPA Epps-Pulley regularizer which approximates a sliced maximum mean discrepancy (MMD) with a Gaussian prior and Gaussian kernel. By expanding the class of viable kernels and priors and computing the closed-form high-dimensional limit of sliced MMDs, we develop alternative KerJEPAs with a number of favorable properties including improved training stability and design flexibility.
PRISM2: Unlocking Multi-Modal General Pathology AI with Clinical Dialogue
Jun 16, 2025Abstract:Recent pathology foundation models can provide rich tile-level representations but fall short of delivering general-purpose clinical utility without further extensive model development. These models lack whole-slide image (WSI) understanding and are not trained with large-scale diagnostic data, limiting their performance on diverse downstream tasks. We introduce PRISM2, a multi-modal slide-level foundation model trained via clinical dialogue to enable scalable, generalizable pathology AI. PRISM2 is trained on nearly 700,000 specimens (2.3 million WSIs) paired with real-world clinical diagnostic reports in a two-stage process. In Stage 1, a vision-language model is trained using contrastive and captioning objectives to align whole slide embeddings with textual clinical diagnosis. In Stage 2, the language model is unfrozen to enable diagnostic conversation and extract more clinically meaningful representations from hidden states. PRISM2 achieves strong performance on diagnostic and biomarker prediction tasks, outperforming prior slide-level models including PRISM and TITAN. It also introduces a zero-shot yes/no classification approach that surpasses CLIP-style methods without prompt tuning or class enumeration. By aligning visual features with clinical reasoning, PRISM2 improves generalization on both data-rich and low-sample tasks, offering a scalable path forward for building general pathology AI agents capable of assisting diagnostic and prognostic decisions.
Virchow2: Scaling Self-Supervised Mixed Magnification Models in Pathology
Aug 14, 2024Abstract:Foundation models are rapidly being developed for computational pathology applications. However, it remains an open question which factors are most important for downstream performance with data scale and diversity, model size, and training algorithm all playing a role. In this work, we propose algorithmic modifications, tailored for pathology, and we present the result of scaling both data and model size, surpassing previous studies in both dimensions. We introduce two new models: Virchow2, a 632 million parameter vision transformer, and Virchow2G, a 1.9 billion parameter vision transformer, each trained with 3.1 million histopathology whole slide images, with diverse tissues, originating institutions, and stains. We achieve state of the art performance on 12 tile-level tasks, as compared to the top performing competing models. Our results suggest that data diversity and domain-specific methods can outperform models that only scale in the number of parameters, but, on average, performance benefits from the combination of domain-specific methods, data scale, and model scale.
Virchow 2: Scaling Self-Supervised Mixed Magnification Models in Pathology
Aug 01, 2024Abstract:Foundation models are rapidly being developed for computational pathology applications. However, it remains an open question which factors are most important for downstream performance with data scale and diversity, model size, and training algorithm all playing a role. In this work, we present the result of scaling both data and model size, surpassing previous studies in both dimensions, and introduce two new models: Virchow 2, a 632M parameter vision transformer, and Virchow 2G, a 1.85B parameter vision transformer, each trained with 3.1M histopathology whole slide images. To support this scale, we propose domain-inspired adaptations to the DINOv2 training algorithm, which is quickly becoming the default method in self-supervised learning for computational pathology. We achieve state of the art performance on twelve tile-level tasks, as compared to the top performing competing models. Our results suggest that data diversity and domain-specific training can outperform models that only scale in the number of parameters, but, on average, performance benefits from domain-tailoring, data scale, and model scale.
PRISM: A Multi-Modal Generative Foundation Model for Slide-Level Histopathology
May 16, 2024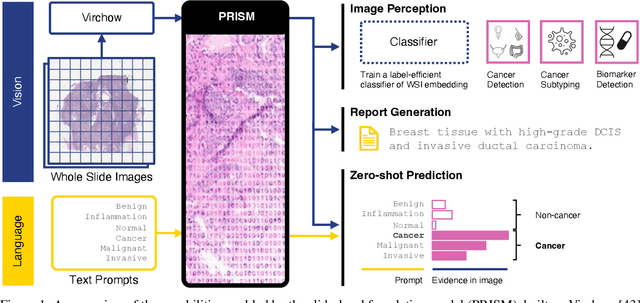
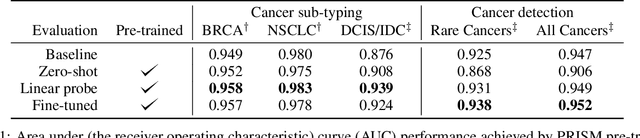
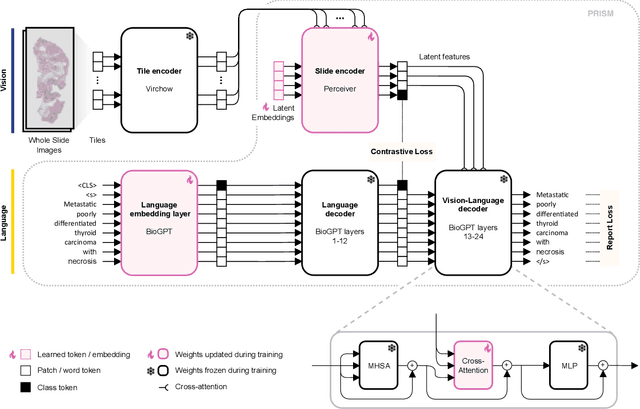
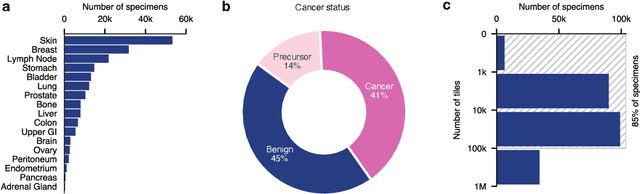
Abstract:Foundation models in computational pathology promise to unlock the development of new clinical decision support systems and models for precision medicine. However, there is a mismatch between most clinical analysis, which is defined at the level of one or more whole slide images, and foundation models to date, which process the thousands of image tiles contained in a whole slide image separately. The requirement to train a network to aggregate information across a large number of tiles in multiple whole slide images limits these models' impact. In this work, we present a slide-level foundation model for H&E-stained histopathology, PRISM, that builds on Virchow tile embeddings and leverages clinical report text for pre-training. Using the tile embeddings, PRISM produces slide-level embeddings with the ability to generate clinical reports, resulting in several modes of use. Using text prompts, PRISM achieves zero-shot cancer detection and sub-typing performance approaching and surpassing that of a supervised aggregator model. Using the slide embeddings with linear classifiers, PRISM surpasses supervised aggregator models. Furthermore, we demonstrate that fine-tuning of the PRISM slide encoder yields label-efficient training for biomarker prediction, a task that typically suffers from low availability of training data; an aggregator initialized with PRISM and trained on as little as 10% of the training data can outperform a supervised baseline that uses all of the data.
Adapting Self-Supervised Learning for Computational Pathology
May 02, 2024Abstract:Self-supervised learning (SSL) has emerged as a key technique for training networks that can generalize well to diverse tasks without task-specific supervision. This property makes SSL desirable for computational pathology, the study of digitized images of tissues, as there are many target applications and often limited labeled training samples. However, SSL algorithms and models have been primarily developed in the field of natural images and whether their performance can be improved by adaptation to particular domains remains an open question. In this work, we present an investigation of modifications to SSL for pathology data, specifically focusing on the DINOv2 algorithm. We propose alternative augmentations, regularization functions, and position encodings motivated by the characteristics of pathology images. We evaluate the impact of these changes on several benchmarks to demonstrate the value of tailored approaches.
Benchmarking a Benchmark: How Reliable is MS-COCO?
Nov 05, 2023Abstract:Benchmark datasets are used to profile and compare algorithms across a variety of tasks, ranging from image classification to segmentation, and also play a large role in image pretraining algorithms. Emphasis is placed on results with little regard to the actual content within the dataset. It is important to question what kind of information is being learned from these datasets and what are the nuances and biases within them. In the following work, Sama-COCO, a re-annotation of MS-COCO, is used to discover potential biases by leveraging a shape analysis pipeline. A model is trained and evaluated on both datasets to examine the impact of different annotation conditions. Results demonstrate that annotation styles are important and that annotation pipelines should closely consider the task of interest. The dataset is made publicly available at https://www.sama.com/sama-coco-dataset/ .
An Empirical Study of Uncertainty in Polygon Annotation and the Impact of Quality Assurance
Nov 05, 2023



Abstract:Polygons are a common annotation format used for quickly annotating objects in instance segmentation tasks. However, many real-world annotation projects request near pixel-perfect labels. While strict pixel guidelines may appear to be the solution to a successful project, practitioners often fail to assess the feasibility of the work requested, and overlook common factors that may challenge the notion of quality. This paper aims to examine and quantify the inherent uncertainty for polygon annotations and the role that quality assurance plays in minimizing its effect. To this end, we conduct an analysis on multi-rater polygon annotations for several objects from the MS-COCO dataset. The results demonstrate that the reliability of a polygon annotation is dependent on a reviewing procedure, as well as the scene and shape complexity.
Surface Denoising based on Normal Filtering in a Robust Statistics Framework
Jul 02, 2020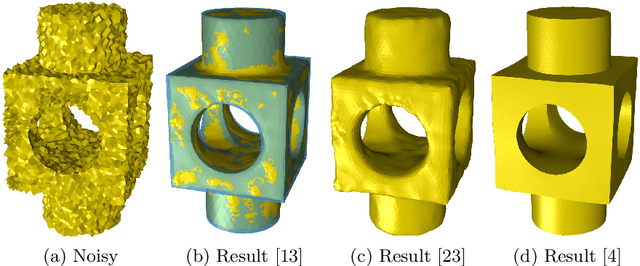
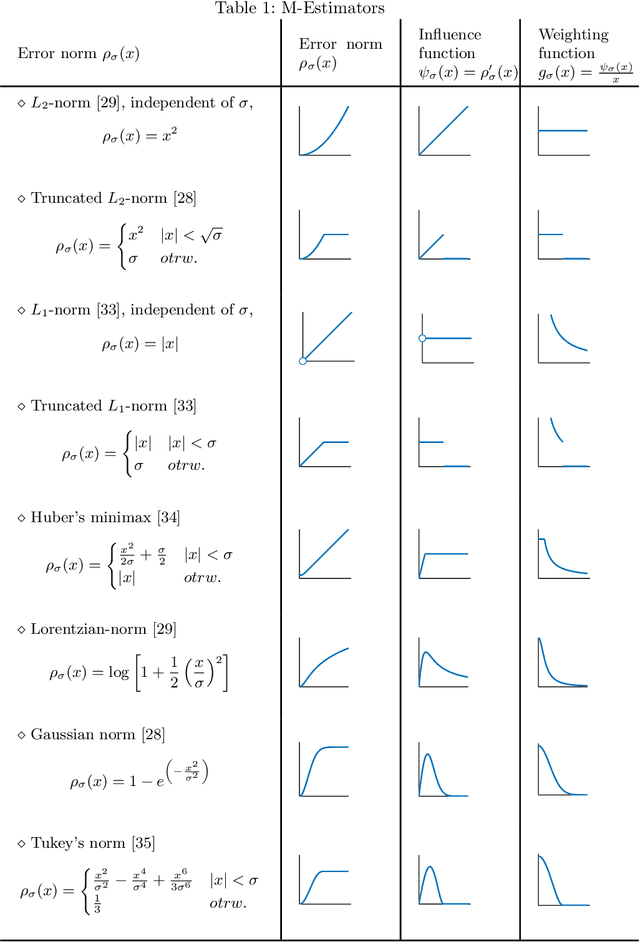
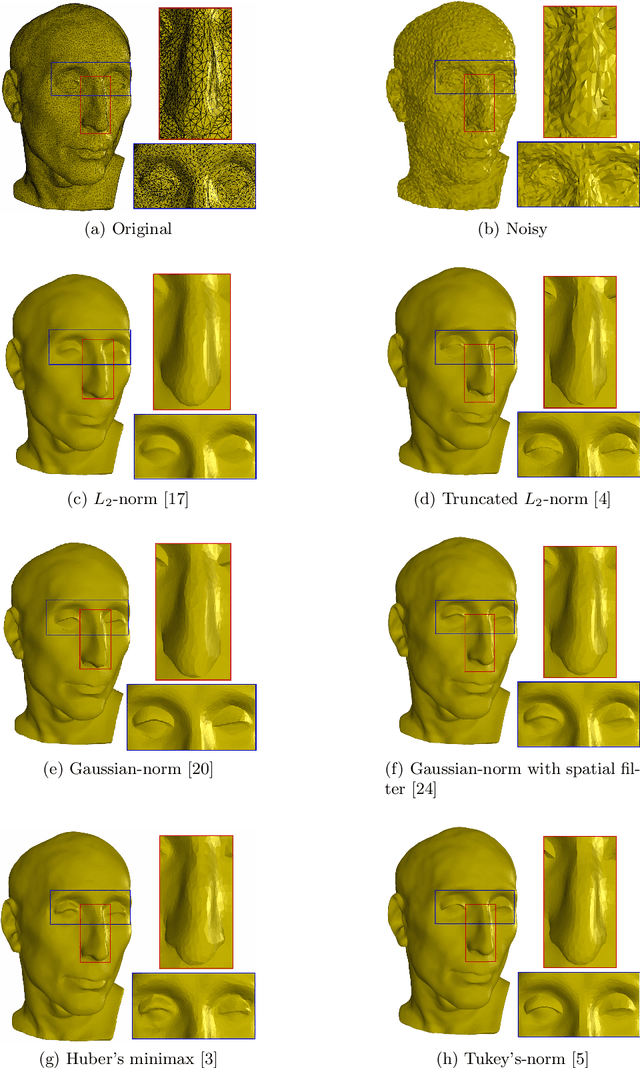
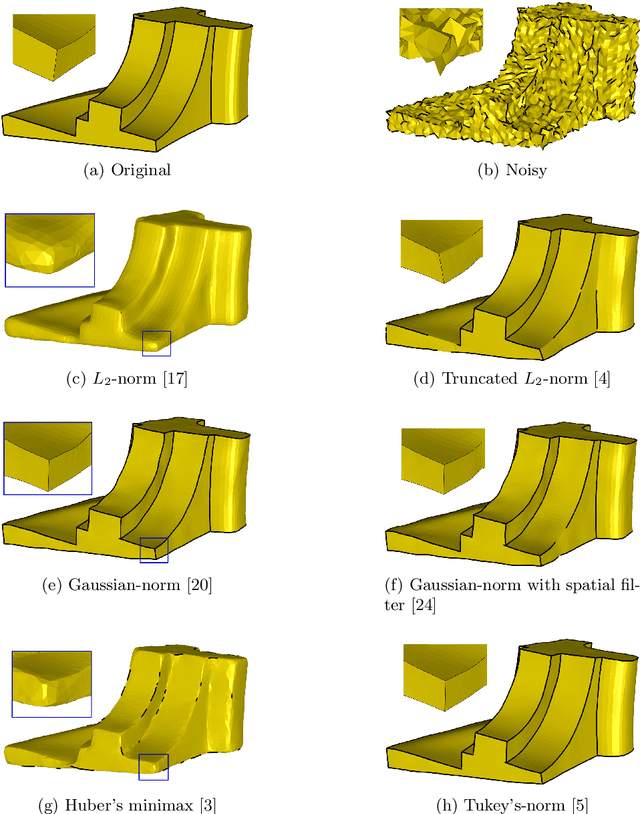
Abstract:During a surface acquisition process using 3D scanners, noise is inevitable and an important step in geometry processing is to remove these noise components from these surfaces (given as points-set or triangulated mesh). The noise-removal process (denoising) can be performed by filtering the surface normals first and by adjusting the vertex positions according to filtered normals afterwards. Therefore, in many available denoising algorithms, the computation of noise-free normals is a key factor. A variety of filters have been introduced for noise-removal from normals, with different focus points like robustness against outliers or large amplitude of noise. Although these filters are performing well in different aspects, a unified framework is missing to establish the relation between them and to provide a theoretical analysis beyond the performance of each method. In this paper, we introduce such a framework to establish relations between a number of widely-used nonlinear filters for face normals in mesh denoising and vertex normals in point set denoising. We cover robust statistical estimation with M-smoothers and their application to linear and non-linear normal filtering. Although these methods originate in different mathematical theories - which include diffusion-, bilateral-, and directional curvature-based algorithms - we demonstrate that all of them can be cast into a unified framework of robust statistics using robust error norms and their corresponding influence functions. This unification contributes to a better understanding of the individual methods and their relations with each other. Furthermore, the presented framework provides a platform for new techniques to combine the advantages of known filters and to compare them with available methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge