Kristen Severson
PRISM2: Unlocking Multi-Modal General Pathology AI with Clinical Dialogue
Jun 16, 2025Abstract:Recent pathology foundation models can provide rich tile-level representations but fall short of delivering general-purpose clinical utility without further extensive model development. These models lack whole-slide image (WSI) understanding and are not trained with large-scale diagnostic data, limiting their performance on diverse downstream tasks. We introduce PRISM2, a multi-modal slide-level foundation model trained via clinical dialogue to enable scalable, generalizable pathology AI. PRISM2 is trained on nearly 700,000 specimens (2.3 million WSIs) paired with real-world clinical diagnostic reports in a two-stage process. In Stage 1, a vision-language model is trained using contrastive and captioning objectives to align whole slide embeddings with textual clinical diagnosis. In Stage 2, the language model is unfrozen to enable diagnostic conversation and extract more clinically meaningful representations from hidden states. PRISM2 achieves strong performance on diagnostic and biomarker prediction tasks, outperforming prior slide-level models including PRISM and TITAN. It also introduces a zero-shot yes/no classification approach that surpasses CLIP-style methods without prompt tuning or class enumeration. By aligning visual features with clinical reasoning, PRISM2 improves generalization on both data-rich and low-sample tasks, offering a scalable path forward for building general pathology AI agents capable of assisting diagnostic and prognostic decisions.
Virchow2: Scaling Self-Supervised Mixed Magnification Models in Pathology
Aug 14, 2024Abstract:Foundation models are rapidly being developed for computational pathology applications. However, it remains an open question which factors are most important for downstream performance with data scale and diversity, model size, and training algorithm all playing a role. In this work, we propose algorithmic modifications, tailored for pathology, and we present the result of scaling both data and model size, surpassing previous studies in both dimensions. We introduce two new models: Virchow2, a 632 million parameter vision transformer, and Virchow2G, a 1.9 billion parameter vision transformer, each trained with 3.1 million histopathology whole slide images, with diverse tissues, originating institutions, and stains. We achieve state of the art performance on 12 tile-level tasks, as compared to the top performing competing models. Our results suggest that data diversity and domain-specific methods can outperform models that only scale in the number of parameters, but, on average, performance benefits from the combination of domain-specific methods, data scale, and model scale.
Virchow 2: Scaling Self-Supervised Mixed Magnification Models in Pathology
Aug 01, 2024Abstract:Foundation models are rapidly being developed for computational pathology applications. However, it remains an open question which factors are most important for downstream performance with data scale and diversity, model size, and training algorithm all playing a role. In this work, we present the result of scaling both data and model size, surpassing previous studies in both dimensions, and introduce two new models: Virchow 2, a 632M parameter vision transformer, and Virchow 2G, a 1.85B parameter vision transformer, each trained with 3.1M histopathology whole slide images. To support this scale, we propose domain-inspired adaptations to the DINOv2 training algorithm, which is quickly becoming the default method in self-supervised learning for computational pathology. We achieve state of the art performance on twelve tile-level tasks, as compared to the top performing competing models. Our results suggest that data diversity and domain-specific training can outperform models that only scale in the number of parameters, but, on average, performance benefits from domain-tailoring, data scale, and model scale.
PRISM: A Multi-Modal Generative Foundation Model for Slide-Level Histopathology
May 16, 2024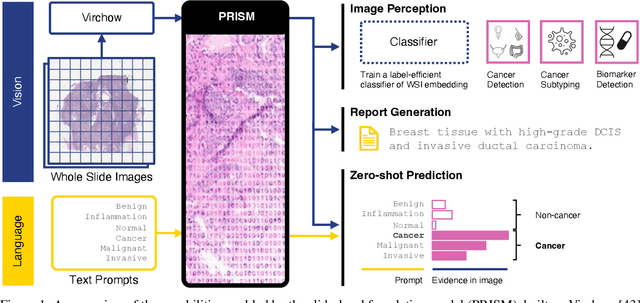
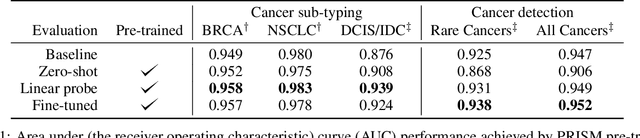
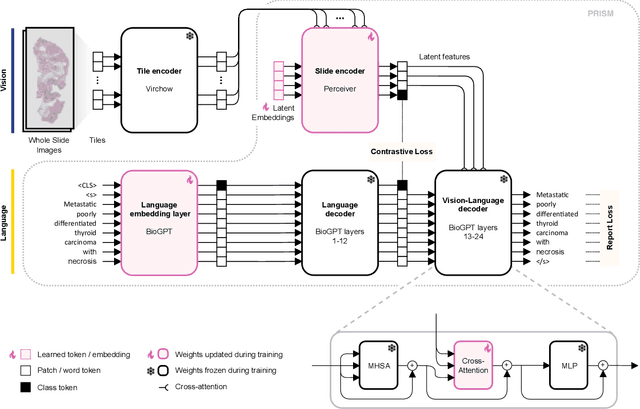
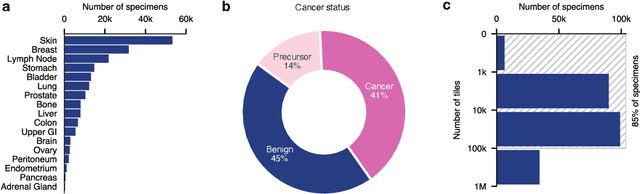
Abstract:Foundation models in computational pathology promise to unlock the development of new clinical decision support systems and models for precision medicine. However, there is a mismatch between most clinical analysis, which is defined at the level of one or more whole slide images, and foundation models to date, which process the thousands of image tiles contained in a whole slide image separately. The requirement to train a network to aggregate information across a large number of tiles in multiple whole slide images limits these models' impact. In this work, we present a slide-level foundation model for H&E-stained histopathology, PRISM, that builds on Virchow tile embeddings and leverages clinical report text for pre-training. Using the tile embeddings, PRISM produces slide-level embeddings with the ability to generate clinical reports, resulting in several modes of use. Using text prompts, PRISM achieves zero-shot cancer detection and sub-typing performance approaching and surpassing that of a supervised aggregator model. Using the slide embeddings with linear classifiers, PRISM surpasses supervised aggregator models. Furthermore, we demonstrate that fine-tuning of the PRISM slide encoder yields label-efficient training for biomarker prediction, a task that typically suffers from low availability of training data; an aggregator initialized with PRISM and trained on as little as 10% of the training data can outperform a supervised baseline that uses all of the data.
Adapting Self-Supervised Learning for Computational Pathology
May 02, 2024Abstract:Self-supervised learning (SSL) has emerged as a key technique for training networks that can generalize well to diverse tasks without task-specific supervision. This property makes SSL desirable for computational pathology, the study of digitized images of tissues, as there are many target applications and often limited labeled training samples. However, SSL algorithms and models have been primarily developed in the field of natural images and whether their performance can be improved by adaptation to particular domains remains an open question. In this work, we present an investigation of modifications to SSL for pathology data, specifically focusing on the DINOv2 algorithm. We propose alternative augmentations, regularization functions, and position encodings motivated by the characteristics of pathology images. We evaluate the impact of these changes on several benchmarks to demonstrate the value of tailored approaches.
Improving mitosis detection on histopathology images using large vision-language models
Oct 11, 2023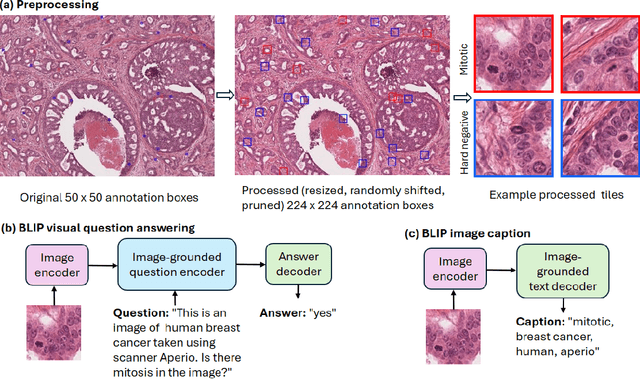
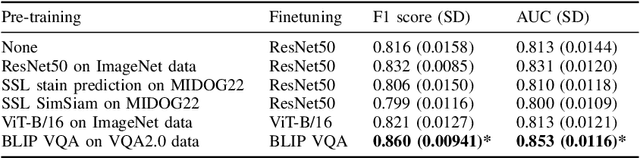
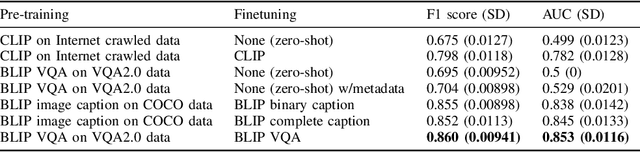
Abstract:In certain types of cancerous tissue, mitotic count has been shown to be associated with tumor proliferation, poor prognosis, and therapeutic resistance. Due to the high inter-rater variability of mitotic counting by pathologists, convolutional neural networks (CNNs) have been employed to reduce the subjectivity of mitosis detection in hematoxylin and eosin (H&E)-stained whole slide images. However, most existing models have performance that lags behind expert panel review and only incorporate visual information. In this work, we demonstrate that pre-trained large-scale vision-language models that leverage both visual features and natural language improve mitosis detection accuracy. We formulate the mitosis detection task as an image captioning task and a visual question answering (VQA) task by including metadata such as tumor and scanner types as context. The effectiveness of our pipeline is demonstrated via comparison with various baseline models using 9,501 mitotic figures and 11,051 hard negatives (non-mitotic figures that are difficult to characterize) from the publicly available Mitosis Domain Generalization Challenge (MIDOG22) dataset.
Unsupervised learning with contrastive latent variable models
Nov 14, 2018



Abstract:In unsupervised learning, dimensionality reduction is an important tool for data exploration and visualization. Because these aims are typically open-ended, it can be useful to frame the problem as looking for patterns that are enriched in one dataset relative to another. These pairs of datasets occur commonly, for instance a population of interest vs. control or signal vs. signal free recordings.However, there are few methods that work on sets of data as opposed to data points or sequences. Here, we present a probabilistic model for dimensionality reduction to discover signal that is enriched in the target dataset relative to the background dataset. The data in these sets do not need to be paired or grouped beyond set membership. By using a probabilistic model where some structure is shared amongst the two datasets and some is unique to the target dataset, we are able to recover interesting structure in the latent space of the target dataset. The method also has the advantages of a probabilistic model, namely that it allows for the incorporation of prior information, handles missing data, and can be generalized to different distributional assumptions. We describe several possible variations of the model and demonstrate the application of the technique to de-noising, feature selection, and subgroup discovery settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge