Ruiwen Ding
Semi-Supervised Domain Adaptation with Latent Diffusion for Pathology Image Classification
Jan 23, 2026Abstract:Deep learning models in computational pathology often fail to generalize across cohorts and institutions due to domain shift. Existing approaches either fail to leverage unlabeled data from the target domain or rely on image-to-image translation, which can distort tissue structures and compromise model accuracy. In this work, we propose a semi-supervised domain adaptation (SSDA) framework that utilizes a latent diffusion model trained on unlabeled data from both the source and target domains to generate morphology-preserving and target-aware synthetic images. By conditioning the diffusion model on foundation model features, cohort identity, and tissue preparation method, we preserve tissue structure in the source domain while introducing target-domain appearance characteristics. The target-aware synthetic images, combined with real, labeled images from the source cohort, are subsequently used to train a downstream classifier, which is then tested on the target cohort. The effectiveness of the proposed SSDA framework is demonstrated on the task of lung adenocarcinoma prognostication. The proposed augmentation yielded substantially better performance on the held-out test set from the target cohort, without degrading source-cohort performance. The approach improved the weighted F1 score on the target-cohort held-out test set from 0.611 to 0.706 and the macro F1 score from 0.641 to 0.716. Our results demonstrate that target-aware diffusion-based synthetic data augmentation provides a promising and effective approach for improving domain generalization in computational pathology.
Predicting ulcer in H&E images of inflammatory bowel disease using domain-knowledge-driven graph neural network
Apr 13, 2025Abstract:Inflammatory bowel disease (IBD) involves chronic inflammation of the digestive tract, with treatment options often burdened by adverse effects. Identifying biomarkers for personalized treatment is crucial. While immune cells play a key role in IBD, accurately identifying ulcer regions in whole slide images (WSIs) is essential for characterizing these cells and exploring potential therapeutics. Multiple instance learning (MIL) approaches have advanced WSI analysis but they lack spatial context awareness. In this work, we propose a weakly-supervised model called DomainGCN that employs a graph convolution neural network (GCN) and incorporates domain-specific knowledge of ulcer features, specifically, the presence of epithelium, lymphocytes, and debris for WSI-level ulcer prediction in IBD. We demonstrate that DomainGCN outperforms various state-of-the-art (SOTA) MIL methods and show the added value of domain knowledge.
Combining Graph Neural Network and Mamba to Capture Local and Global Tissue Spatial Relationships in Whole Slide Images
Jun 05, 2024Abstract:In computational pathology, extracting spatial features from gigapixel whole slide images (WSIs) is a fundamental task, but due to their large size, WSIs are typically segmented into smaller tiles. A critical aspect of this analysis is aggregating information from these tiles to make predictions at the WSI level. We introduce a model that combines a message-passing graph neural network (GNN) with a state space model (Mamba) to capture both local and global spatial relationships among the tiles in WSIs. The model's effectiveness was demonstrated in predicting progression-free survival among patients with early-stage lung adenocarcinomas (LUAD). We compared the model with other state-of-the-art methods for tile-level information aggregation in WSIs, including tile-level information summary statistics-based aggregation, multiple instance learning (MIL)-based aggregation, GNN-based aggregation, and GNN-transformer-based aggregation. Additional experiments showed the impact of different types of node features and different tile sampling strategies on the model performance. This work can be easily extended to any WSI-based analysis. Code: https://github.com/rina-ding/gat-mamba.
Improving mitosis detection on histopathology images using large vision-language models
Oct 11, 2023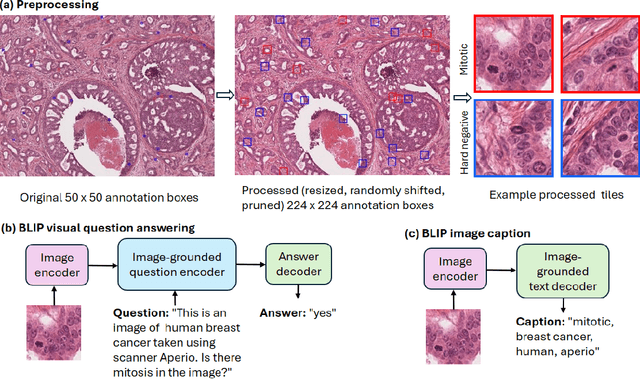
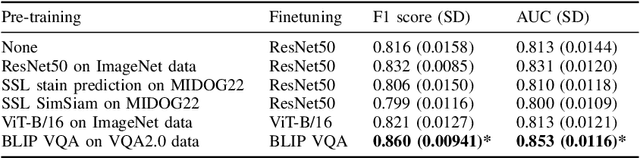
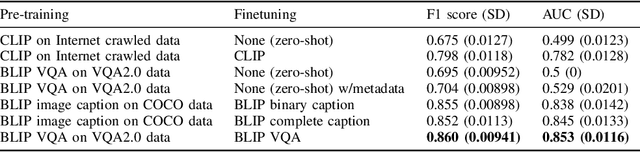
Abstract:In certain types of cancerous tissue, mitotic count has been shown to be associated with tumor proliferation, poor prognosis, and therapeutic resistance. Due to the high inter-rater variability of mitotic counting by pathologists, convolutional neural networks (CNNs) have been employed to reduce the subjectivity of mitosis detection in hematoxylin and eosin (H&E)-stained whole slide images. However, most existing models have performance that lags behind expert panel review and only incorporate visual information. In this work, we demonstrate that pre-trained large-scale vision-language models that leverage both visual features and natural language improve mitosis detection accuracy. We formulate the mitosis detection task as an image captioning task and a visual question answering (VQA) task by including metadata such as tumor and scanner types as context. The effectiveness of our pipeline is demonstrated via comparison with various baseline models using 9,501 mitotic figures and 11,051 hard negatives (non-mitotic figures that are difficult to characterize) from the publicly available Mitosis Domain Generalization Challenge (MIDOG22) dataset.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge