Kha-Dinh Luong
Global Human-guided Counterfactual Explanations for Molecular Properties via Reinforcement Learning
Jun 19, 2024Abstract:Counterfactual explanations of Graph Neural Networks (GNNs) offer a powerful way to understand data that can naturally be represented by a graph structure. Furthermore, in many domains, it is highly desirable to derive data-driven global explanations or rules that can better explain the high-level properties of the models and data in question. However, evaluating global counterfactual explanations is hard in real-world datasets due to a lack of human-annotated ground truth, which limits their use in areas like molecular sciences. Additionally, the increasing scale of these datasets provides a challenge for random search-based methods. In this paper, we develop a novel global explanation model RLHEX for molecular property prediction. It aligns the counterfactual explanations with human-defined principles, making the explanations more interpretable and easy for experts to evaluate. RLHEX includes a VAE-based graph generator to generate global explanations and an adapter to adjust the latent representation space to human-defined principles. Optimized by Proximal Policy Optimization (PPO), the global explanations produced by RLHEX cover 4.12% more input graphs and reduce the distance between the counterfactual explanation set and the input set by 0.47% on average across three molecular datasets. RLHEX provides a flexible framework to incorporate different human-designed principles into the counterfactual explanation generation process, aligning these explanations with domain expertise. The code and data are released at https://github.com/dqwang122/RLHEX.
Combining Graph Neural Network and Mamba to Capture Local and Global Tissue Spatial Relationships in Whole Slide Images
Jun 05, 2024Abstract:In computational pathology, extracting spatial features from gigapixel whole slide images (WSIs) is a fundamental task, but due to their large size, WSIs are typically segmented into smaller tiles. A critical aspect of this analysis is aggregating information from these tiles to make predictions at the WSI level. We introduce a model that combines a message-passing graph neural network (GNN) with a state space model (Mamba) to capture both local and global spatial relationships among the tiles in WSIs. The model's effectiveness was demonstrated in predicting progression-free survival among patients with early-stage lung adenocarcinomas (LUAD). We compared the model with other state-of-the-art methods for tile-level information aggregation in WSIs, including tile-level information summary statistics-based aggregation, multiple instance learning (MIL)-based aggregation, GNN-based aggregation, and GNN-transformer-based aggregation. Additional experiments showed the impact of different types of node features and different tile sampling strategies on the model performance. This work can be easily extended to any WSI-based analysis. Code: https://github.com/rina-ding/gat-mamba.
Fragment-based Pretraining and Finetuning on Molecular Graphs
Oct 05, 2023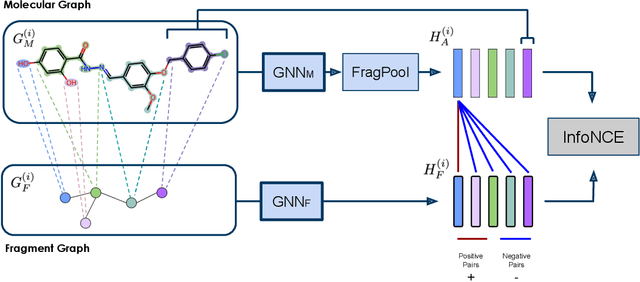
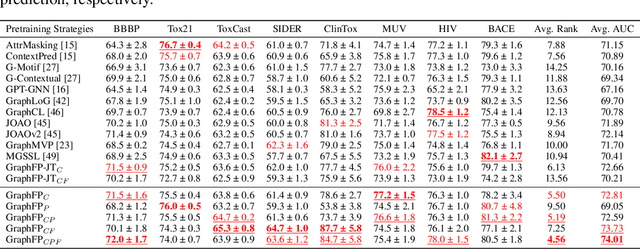
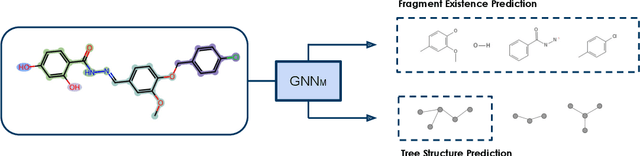
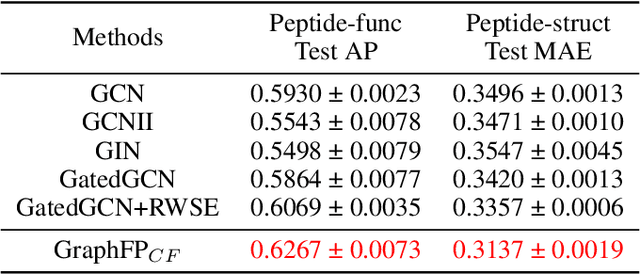
Abstract:Property prediction on molecular graphs is an important application of Graph Neural Networks (GNNs). Recently, unlabeled molecular data has become abundant, which facilitates the rapid development of self-supervised learning for GNNs in the chemical domain. In this work, we propose pretraining GNNs at the fragment level, which serves as a promising middle ground to overcome the limitations of node-level and graph-level pretraining. Borrowing techniques from recent work on principle subgraph mining, we obtain a compact vocabulary of prevalent fragments that span a large pretraining dataset. From the extracted vocabulary, we introduce several fragment-based contrastive and predictive pretraining tasks. The contrastive learning task jointly pretrains two different GNNs: one based on molecular graphs and one based on fragment graphs, which represents high-order connectivity within molecules. By enforcing the consistency between the fragment embedding and the aggregated embedding of the corresponding atoms from the molecular graphs, we ensure that both embeddings capture structural information at multiple resolutions. The structural information of the fragment graphs is further exploited to extract auxiliary labels for the graph-level predictive pretraining. We employ both the pretrained molecular-based and fragment-based GNNs for downstream prediction, thus utilizing the fragment information during finetuning. Our models advance the performances on 5 out of 8 common molecular benchmarks and improve the performances on long-range biological benchmarks by at least 11.5%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge