Ambuj Singh
FlowSymm: Physics Aware, Symmetry Preserving Graph Attention for Network Flow Completion
Jan 29, 2026Abstract:Recovering missing flows on the edges of a network, while exactly respecting local conservation laws, is a fundamental inverse problem that arises in many systems such as transportation, energy, and mobility. We introduce FlowSymm, a novel architecture that combines (i) a group-action on divergence-free flows, (ii) a graph-attention encoder to learn feature-conditioned weights over these symmetry-preserving actions, and (iii) a lightweight Tikhonov refinement solved via implicit bilevel optimization. The method first anchors the given observation on a minimum-norm divergence-free completion. We then compute an orthonormal basis for all admissible group actions that leave the observed flows invariant and parameterize the valid solution subspace, which shows an Abelian group structure under vector addition. A stack of GATv2 layers then encodes the graph and its edge features into per-edge embeddings, which are pooled over the missing edges and produce per-basis attention weights. This attention-guided process selects a set of physics-aware group actions that preserve the observed flows. Finally, a scalar Tikhonov penalty refines the missing entries via a convex least-squares solver, with gradients propagated implicitly through Cholesky factorization. Across three real-world flow benchmarks (traffic, power, bike), FlowSymm outperforms state-of-the-art baselines in RMSE, MAE and correlation metrics.
APEX: Empowering LLMs with Physics-Based Task Planning for Real-time Insight
May 20, 2025Abstract:Large Language Models (LLMs) demonstrate strong reasoning and task planning capabilities but remain fundamentally limited in physical interaction modeling. Existing approaches integrate perception via Vision-Language Models (VLMs) or adaptive decision-making through Reinforcement Learning (RL), but they fail to capture dynamic object interactions or require task-specific training, limiting their real-world applicability. We introduce APEX (Anticipatory Physics-Enhanced Execution), a framework that equips LLMs with physics-driven foresight for real-time task planning. APEX constructs structured graphs to identify and model the most relevant dynamic interactions in the environment, providing LLMs with explicit physical state updates. Simultaneously, APEX provides low-latency forward simulations of physically feasible actions, allowing LLMs to select optimal strategies based on predictive outcomes rather than static observations. We evaluate APEX on three benchmarks designed to assess perception, prediction, and decision-making: (1) Physics Reasoning Benchmark, testing causal inference and object motion prediction; (2) Tetris, evaluating whether physics-informed prediction enhances decision-making performance in long-horizon planning tasks; (3) Dynamic Obstacle Avoidance, assessing the immediate integration of perception and action feasibility analysis. APEX significantly outperforms standard LLMs and VLM-based models, demonstrating the necessity of explicit physics reasoning for bridging the gap between language-based intelligence and real-world task execution. The source code and experiment setup are publicly available at https://github.com/hwj20/APEX_EXP .
Efficient Multi Subject Visual Reconstruction from fMRI Using Aligned Representations
May 03, 2025



Abstract:This work introduces a novel approach to fMRI-based visual image reconstruction using a subject-agnostic common representation space. We show that the brain signals of the subjects can be aligned in this common space during training to form a semantically aligned common brain. This is leveraged to demonstrate that aligning subject-specific lightweight modules to a reference subject is significantly more efficient than traditional end-to-end training methods. Our approach excels in low-data scenarios. We evaluate our methods on different datasets, demonstrating that the common space is subject and dataset-agnostic.
Attribute-Enhanced Similarity Ranking for Sparse Link Prediction
Nov 29, 2024



Abstract:Link prediction is a fundamental problem in graph data. In its most realistic setting, the problem consists of predicting missing or future links between random pairs of nodes from the set of disconnected pairs. Graph Neural Networks (GNNs) have become the predominant framework for link prediction. GNN-based methods treat link prediction as a binary classification problem and handle the extreme class imbalance -- real graphs are very sparse -- by sampling (uniformly at random) a balanced number of disconnected pairs not only for training but also for evaluation. However, we show that the reported performance of GNNs for link prediction in the balanced setting does not translate to the more realistic imbalanced setting and that simpler topology-based approaches are often better at handling sparsity. These findings motivate Gelato, a similarity-based link-prediction method that applies (1) graph learning based on node attributes to enhance a topological heuristic, (2) a ranking loss for addressing class imbalance, and (3) a negative sampling scheme that efficiently selects hard training pairs via graph partitioning. Experiments show that Gelato outperforms existing GNN-based alternatives.
Normalized Space Alignment: A Versatile Metric for Representation Analysis
Nov 07, 2024Abstract:We introduce a manifold analysis technique for neural network representations. Normalized Space Alignment (NSA) compares pairwise distances between two point clouds derived from the same source and having the same size, while potentially possessing differing dimensionalities. NSA can act as both an analytical tool and a differentiable loss function, providing a robust means of comparing and aligning representations across different layers and models. It satisfies the criteria necessary for both a similarity metric and a neural network loss function. We showcase NSA's versatility by illustrating its utility as a representation space analysis metric, a structure-preserving loss function, and a robustness analysis tool. NSA is not only computationally efficient but it can also approximate the global structural discrepancy during mini-batching, facilitating its use in a wide variety of neural network training paradigms.
Global Human-guided Counterfactual Explanations for Molecular Properties via Reinforcement Learning
Jun 19, 2024Abstract:Counterfactual explanations of Graph Neural Networks (GNNs) offer a powerful way to understand data that can naturally be represented by a graph structure. Furthermore, in many domains, it is highly desirable to derive data-driven global explanations or rules that can better explain the high-level properties of the models and data in question. However, evaluating global counterfactual explanations is hard in real-world datasets due to a lack of human-annotated ground truth, which limits their use in areas like molecular sciences. Additionally, the increasing scale of these datasets provides a challenge for random search-based methods. In this paper, we develop a novel global explanation model RLHEX for molecular property prediction. It aligns the counterfactual explanations with human-defined principles, making the explanations more interpretable and easy for experts to evaluate. RLHEX includes a VAE-based graph generator to generate global explanations and an adapter to adjust the latent representation space to human-defined principles. Optimized by Proximal Policy Optimization (PPO), the global explanations produced by RLHEX cover 4.12% more input graphs and reduce the distance between the counterfactual explanation set and the input set by 0.47% on average across three molecular datasets. RLHEX provides a flexible framework to incorporate different human-designed principles into the counterfactual explanation generation process, aligning these explanations with domain expertise. The code and data are released at https://github.com/dqwang122/RLHEX.
Learning Neural Contracting Dynamics: Extended Linearization and Global Guarantees
Feb 14, 2024



Abstract:Global stability and robustness guarantees in learned dynamical systems are essential to ensure well-behavedness of the systems in the face of uncertainty. We present Extended Linearized Contracting Dynamics (ELCD), the first neural network-based dynamical system with global contractivity guarantees in arbitrary metrics. The key feature of ELCD is a parametrization of the extended linearization of the nonlinear vector field. In its most basic form, ELCD is guaranteed to be (i) globally exponentially stable, (ii) equilibrium contracting, and (iii) globally contracting with respect to some metric. To allow for contraction with respect to more general metrics in the data space, we train diffeomorphisms between the data space and a latent space and enforce contractivity in the latent space, which ensures global contractivity in the data space. We demonstrate the performance of ELCD on the $2$D, $4$D, and $8$D LASA datasets.
DGCLUSTER: A Neural Framework for Attributed Graph Clustering via Modularity Maximization
Dec 20, 2023



Abstract:Graph clustering is a fundamental and challenging task in the field of graph mining where the objective is to group the nodes into clusters taking into consideration the topology of the graph. It has several applications in diverse domains spanning social network analysis, recommender systems, computer vision, and bioinformatics. In this work, we propose a novel method, DGCluster, which primarily optimizes the modularity objective using graph neural networks and scales linearly with the graph size. Our method does not require the number of clusters to be specified as a part of the input and can also leverage the availability of auxiliary node level information. We extensively test DGCluster on several real-world datasets of varying sizes, across multiple popular cluster quality metrics. Our approach consistently outperforms the state-of-the-art methods, demonstrating significant performance gains in almost all settings.
XplainLLM: A QA Explanation Dataset for Understanding LLM Decision-Making
Nov 15, 2023Abstract:Large Language Models (LLMs) have recently made impressive strides in natural language understanding tasks. Despite their remarkable performance, understanding their decision-making process remains a big challenge. In this paper, we look into bringing some transparency to this process by introducing a new explanation dataset for question answering (QA) tasks that integrates knowledge graphs (KGs) in a novel way. Our dataset includes 12,102 question-answer-explanation (QAE) triples. Each explanation in the dataset links the LLM's reasoning to entities and relations in the KGs. The explanation component includes a why-choose explanation, a why-not-choose explanation, and a set of reason-elements that underlie the LLM's decision. We leverage KGs and graph attention networks (GAT) to find the reason-elements and transform them into why-choose and why-not-choose explanations that are comprehensible to humans. Through quantitative and qualitative evaluations, we demonstrate the potential of our dataset to improve the in-context learning of LLMs, and enhance their interpretability and explainability. Our work contributes to the field of explainable AI by enabling a deeper understanding of the LLMs decision-making process to make them more transparent and thereby, potentially more reliable, to researchers and practitioners alike. Our dataset is available at: https://github.com/chen-zichen/XplainLLM_dataset.git
Fragment-based Pretraining and Finetuning on Molecular Graphs
Oct 05, 2023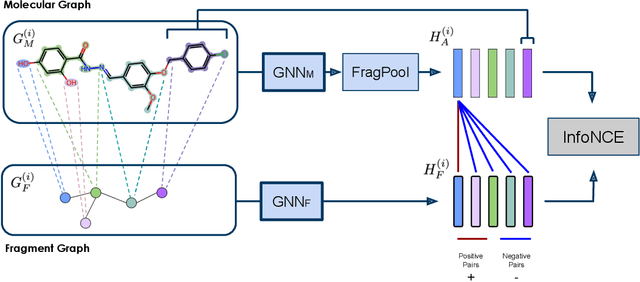
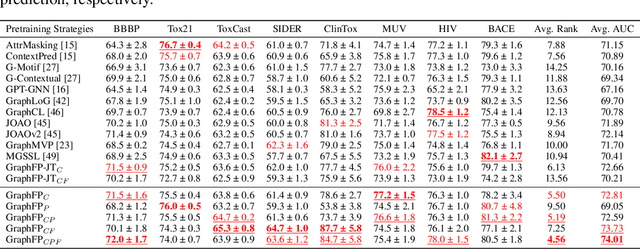
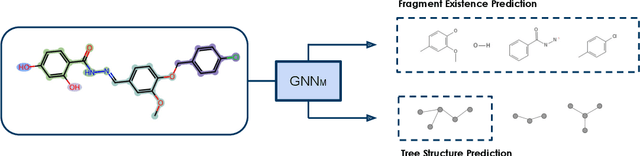
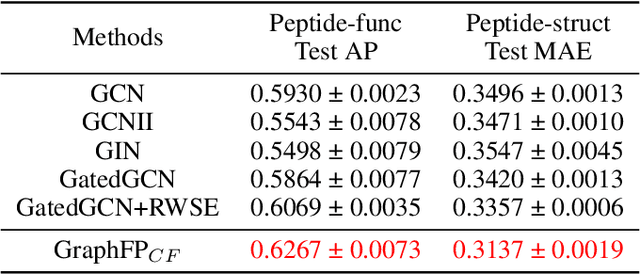
Abstract:Property prediction on molecular graphs is an important application of Graph Neural Networks (GNNs). Recently, unlabeled molecular data has become abundant, which facilitates the rapid development of self-supervised learning for GNNs in the chemical domain. In this work, we propose pretraining GNNs at the fragment level, which serves as a promising middle ground to overcome the limitations of node-level and graph-level pretraining. Borrowing techniques from recent work on principle subgraph mining, we obtain a compact vocabulary of prevalent fragments that span a large pretraining dataset. From the extracted vocabulary, we introduce several fragment-based contrastive and predictive pretraining tasks. The contrastive learning task jointly pretrains two different GNNs: one based on molecular graphs and one based on fragment graphs, which represents high-order connectivity within molecules. By enforcing the consistency between the fragment embedding and the aggregated embedding of the corresponding atoms from the molecular graphs, we ensure that both embeddings capture structural information at multiple resolutions. The structural information of the fragment graphs is further exploited to extract auxiliary labels for the graph-level predictive pretraining. We employ both the pretrained molecular-based and fragment-based GNNs for downstream prediction, thus utilizing the fragment information during finetuning. Our models advance the performances on 5 out of 8 common molecular benchmarks and improve the performances on long-range biological benchmarks by at least 11.5%.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge