Nicolo Fusi
PRISM2: Unlocking Multi-Modal General Pathology AI with Clinical Dialogue
Jun 16, 2025Abstract:Recent pathology foundation models can provide rich tile-level representations but fall short of delivering general-purpose clinical utility without further extensive model development. These models lack whole-slide image (WSI) understanding and are not trained with large-scale diagnostic data, limiting their performance on diverse downstream tasks. We introduce PRISM2, a multi-modal slide-level foundation model trained via clinical dialogue to enable scalable, generalizable pathology AI. PRISM2 is trained on nearly 700,000 specimens (2.3 million WSIs) paired with real-world clinical diagnostic reports in a two-stage process. In Stage 1, a vision-language model is trained using contrastive and captioning objectives to align whole slide embeddings with textual clinical diagnosis. In Stage 2, the language model is unfrozen to enable diagnostic conversation and extract more clinically meaningful representations from hidden states. PRISM2 achieves strong performance on diagnostic and biomarker prediction tasks, outperforming prior slide-level models including PRISM and TITAN. It also introduces a zero-shot yes/no classification approach that surpasses CLIP-style methods without prompt tuning or class enumeration. By aligning visual features with clinical reasoning, PRISM2 improves generalization on both data-rich and low-sample tasks, offering a scalable path forward for building general pathology AI agents capable of assisting diagnostic and prognostic decisions.
IGDA: Interactive Graph Discovery through Large Language Model Agents
Feb 24, 2025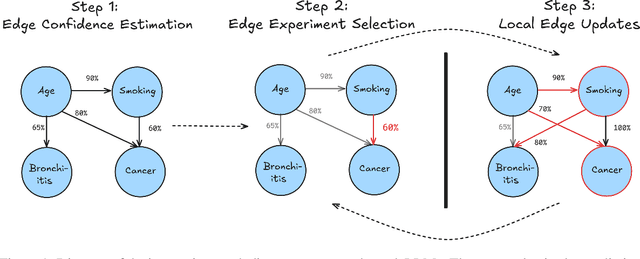
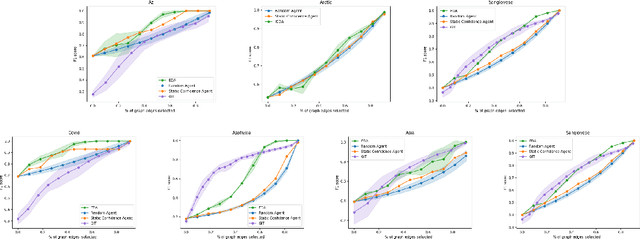
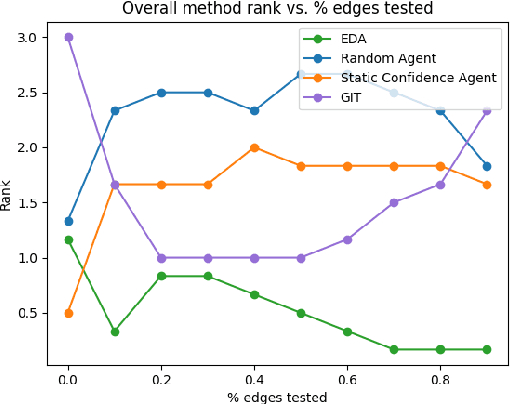
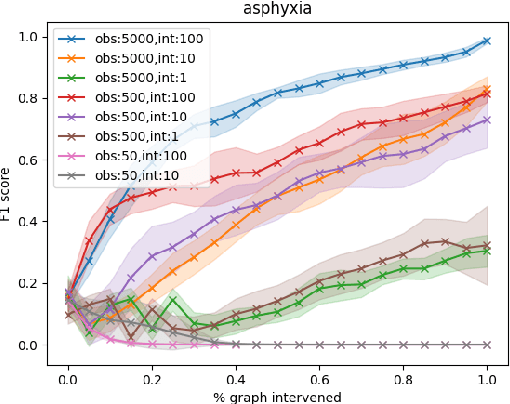
Abstract:Large language models ($\textbf{LLMs}$) have emerged as a powerful method for discovery. Instead of utilizing numerical data, LLMs utilize associated variable $\textit{semantic metadata}$ to predict variable relationships. Simultaneously, LLMs demonstrate impressive abilities to act as black-box optimizers when given an objective $f$ and sequence of trials. We study LLMs at the intersection of these two capabilities by applying LLMs to the task of $\textit{interactive graph discovery}$: given a ground truth graph $G^*$ capturing variable relationships and a budget of $I$ edge experiments over $R$ rounds, minimize the distance between the predicted graph $\hat{G}_R$ and $G^*$ at the end of the $R$-th round. To solve this task we propose $\textbf{IGDA}$, a LLM-based pipeline incorporating two key components: 1) an LLM uncertainty-driven method for edge experiment selection 2) a local graph update strategy utilizing binary feedback from experiments to improve predictions for unselected neighboring edges. Experiments on eight different real-world graphs show our approach often outperforms all baselines including a state-of-the-art numerical method for interactive graph discovery. Further, we conduct a rigorous series of ablations dissecting the impact of each pipeline component. Finally, to assess the impact of memorization, we apply our interactive graph discovery strategy to a complex, new (as of July 2024) causal graph on protein transcription factors, finding strong performance in a setting where memorization is impossible. Overall, our results show IGDA to be a powerful method for graph discovery complementary to existing numerically driven approaches.
Adapting Language Models via Token Translation
Nov 01, 2024Abstract:Modern large language models use a fixed tokenizer to effectively compress text drawn from a source domain. However, applying the same tokenizer to a new target domain often leads to inferior compression, more costly inference, and reduced semantic alignment. To address this deficiency, we introduce Sparse Sinkhorn Token Translation (S2T2). S2T2 trains a tailored tokenizer for the target domain and learns to translate between target and source tokens, enabling more effective reuse of the pre-trained next-source-token predictor. In our experiments with finetuned English language models, S2T2 improves both the perplexity and the compression of out-of-domain protein sequences, outperforming direct finetuning with either the source or target tokenizer. In addition, we find that token translations learned for smaller, less expensive models can be directly transferred to larger, more powerful models to reap the benefits of S2T2 at lower cost.
Virchow2: Scaling Self-Supervised Mixed Magnification Models in Pathology
Aug 14, 2024Abstract:Foundation models are rapidly being developed for computational pathology applications. However, it remains an open question which factors are most important for downstream performance with data scale and diversity, model size, and training algorithm all playing a role. In this work, we propose algorithmic modifications, tailored for pathology, and we present the result of scaling both data and model size, surpassing previous studies in both dimensions. We introduce two new models: Virchow2, a 632 million parameter vision transformer, and Virchow2G, a 1.9 billion parameter vision transformer, each trained with 3.1 million histopathology whole slide images, with diverse tissues, originating institutions, and stains. We achieve state of the art performance on 12 tile-level tasks, as compared to the top performing competing models. Our results suggest that data diversity and domain-specific methods can outperform models that only scale in the number of parameters, but, on average, performance benefits from the combination of domain-specific methods, data scale, and model scale.
Virchow 2: Scaling Self-Supervised Mixed Magnification Models in Pathology
Aug 01, 2024Abstract:Foundation models are rapidly being developed for computational pathology applications. However, it remains an open question which factors are most important for downstream performance with data scale and diversity, model size, and training algorithm all playing a role. In this work, we present the result of scaling both data and model size, surpassing previous studies in both dimensions, and introduce two new models: Virchow 2, a 632M parameter vision transformer, and Virchow 2G, a 1.85B parameter vision transformer, each trained with 3.1M histopathology whole slide images. To support this scale, we propose domain-inspired adaptations to the DINOv2 training algorithm, which is quickly becoming the default method in self-supervised learning for computational pathology. We achieve state of the art performance on twelve tile-level tasks, as compared to the top performing competing models. Our results suggest that data diversity and domain-specific training can outperform models that only scale in the number of parameters, but, on average, performance benefits from domain-tailoring, data scale, and model scale.
PRISM: A Multi-Modal Generative Foundation Model for Slide-Level Histopathology
May 16, 2024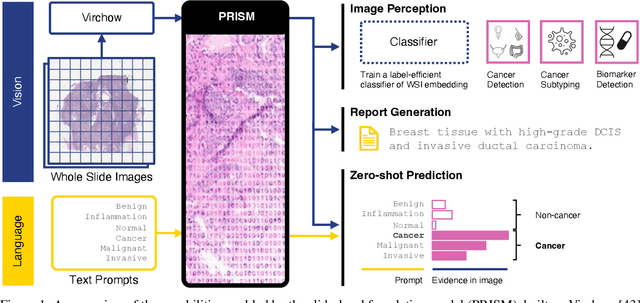
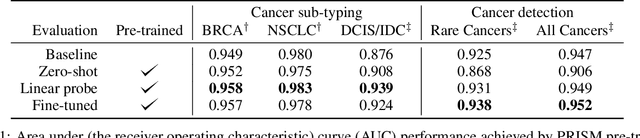
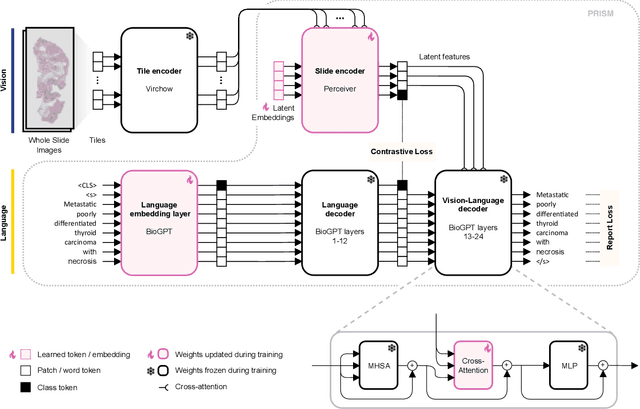
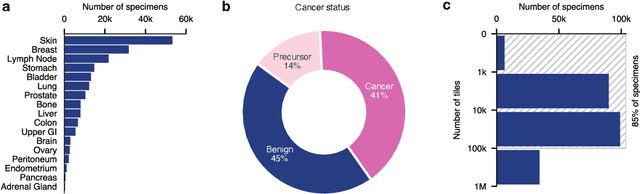
Abstract:Foundation models in computational pathology promise to unlock the development of new clinical decision support systems and models for precision medicine. However, there is a mismatch between most clinical analysis, which is defined at the level of one or more whole slide images, and foundation models to date, which process the thousands of image tiles contained in a whole slide image separately. The requirement to train a network to aggregate information across a large number of tiles in multiple whole slide images limits these models' impact. In this work, we present a slide-level foundation model for H&E-stained histopathology, PRISM, that builds on Virchow tile embeddings and leverages clinical report text for pre-training. Using the tile embeddings, PRISM produces slide-level embeddings with the ability to generate clinical reports, resulting in several modes of use. Using text prompts, PRISM achieves zero-shot cancer detection and sub-typing performance approaching and surpassing that of a supervised aggregator model. Using the slide embeddings with linear classifiers, PRISM surpasses supervised aggregator models. Furthermore, we demonstrate that fine-tuning of the PRISM slide encoder yields label-efficient training for biomarker prediction, a task that typically suffers from low availability of training data; an aggregator initialized with PRISM and trained on as little as 10% of the training data can outperform a supervised baseline that uses all of the data.
Tag-LLM: Repurposing General-Purpose LLMs for Specialized Domains
Feb 06, 2024


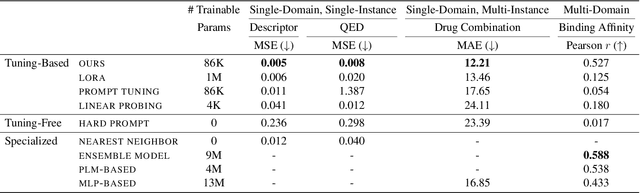
Abstract:Large Language Models (LLMs) have demonstrated remarkable proficiency in understanding and generating natural language. However, their capabilities wane in highly specialized domains underrepresented in the pretraining corpus, such as physical and biomedical sciences. This work explores how to repurpose general LLMs into effective task solvers for specialized domains. We introduce a novel, model-agnostic framework for learning custom input tags, which are parameterized as continuous vectors appended to the LLM's embedding layer, to condition the LLM. We design two types of input tags: domain tags are used to delimit specialized representations (e.g., chemical formulas) and provide domain-relevant context; function tags are used to represent specific functions (e.g., predicting molecular properties) and compress function-solving instructions. We develop a three-stage protocol to learn these tags using auxiliary data and domain knowledge. By explicitly disentangling task domains from task functions, our method enables zero-shot generalization to unseen problems through diverse combinations of the input tags. It also boosts LLM's performance in various specialized domains, such as predicting protein or chemical properties and modeling drug-target interactions, outperforming expert models tailored to these tasks.
Can Generalist Foundation Models Outcompete Special-Purpose Tuning? Case Study in Medicine
Nov 28, 2023
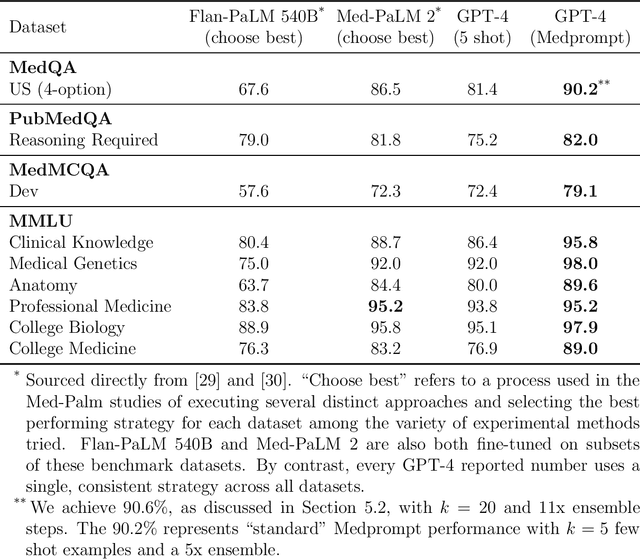

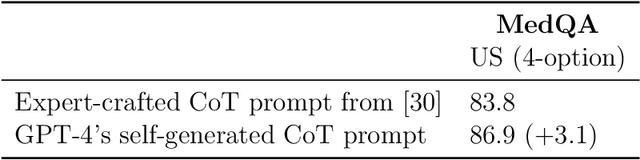
Abstract:Generalist foundation models such as GPT-4 have displayed surprising capabilities in a wide variety of domains and tasks. Yet, there is a prevalent assumption that they cannot match specialist capabilities of fine-tuned models. For example, most explorations to date on medical competency benchmarks have leveraged domain-specific training, as exemplified by efforts on BioGPT and Med-PaLM. We build on a prior study of GPT-4's capabilities on medical challenge benchmarks in the absence of special training. Rather than using simple prompting to highlight the model's out-of-the-box capabilities, we perform a systematic exploration of prompt engineering. We find that prompting innovation can unlock deeper specialist capabilities and show that GPT-4 easily tops prior leading results for medical benchmarks. The prompting methods we explore are general purpose, and make no specific use of domain expertise, removing the need for expert-curated content. Our experimental design carefully controls for overfitting during the prompt engineering process. We introduce Medprompt, based on a composition of several prompting strategies. With Medprompt, GPT-4 achieves state-of-the-art results on all nine of the benchmark datasets in the MultiMedQA suite. The method outperforms leading specialist models such as Med-PaLM 2 by a significant margin with an order of magnitude fewer calls to the model. Steering GPT-4 with Medprompt achieves a 27% reduction in error rate on the MedQA dataset over the best methods to date achieved with specialist models and surpasses a score of 90% for the first time. Beyond medical problems, we show the power of Medprompt to generalize to other domains and provide evidence for the broad applicability of the approach via studies of the strategy on exams in electrical engineering, machine learning, philosophy, accounting, law, nursing, and clinical psychology.
Interpretable Distribution Shift Detection using Optimal Transport
Aug 04, 2022
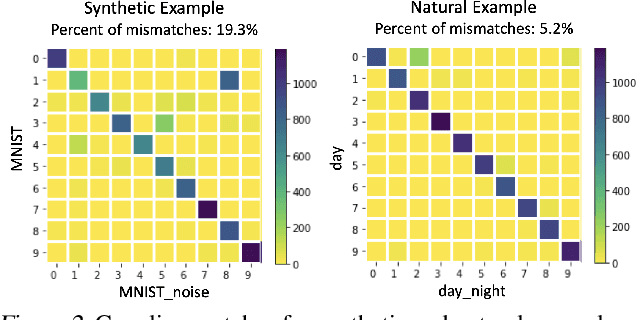
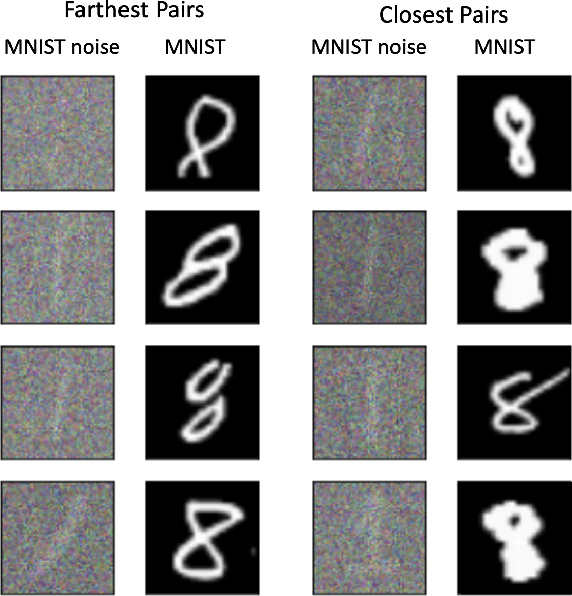

Abstract:We propose a method to identify and characterize distribution shifts in classification datasets based on optimal transport. It allows the user to identify the extent to which each class is affected by the shift, and retrieves corresponding pairs of samples to provide insights on its nature. We illustrate its use on synthetic and natural shift examples. While the results we present are preliminary, we hope that this inspires future work on interpretable methods for analyzing distribution shifts.
Bayesian Optimization Over Iterative Learners with Structured Responses: A Budget-aware Planning Approach
Jun 25, 2022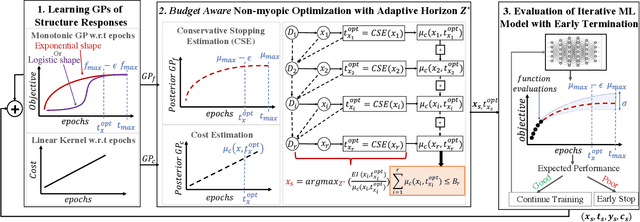
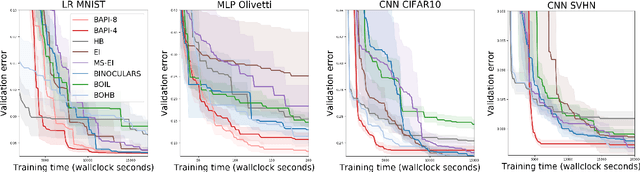
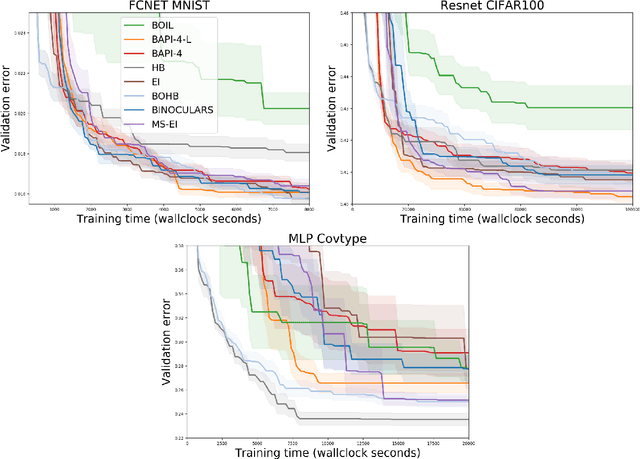
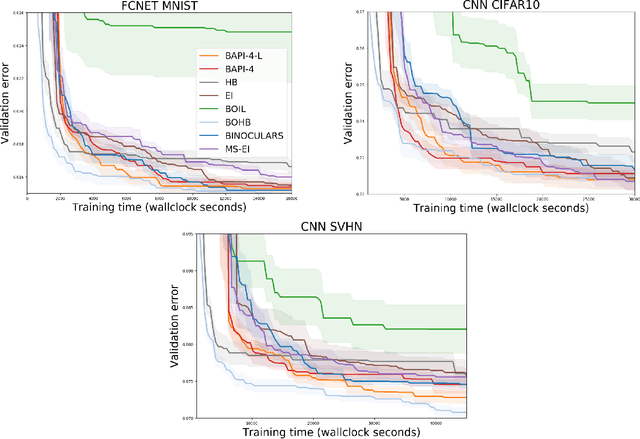
Abstract:The rising growth of deep neural networks (DNNs) and datasets in size motivates the need for efficient solutions for simultaneous model selection and training. Many methods for hyperparameter optimization (HPO) of iterative learners including DNNs attempt to solve this problem by querying and learning a response surface while searching for the optimum of that surface. However, many of these methods make myopic queries, do not consider prior knowledge about the response structure, and/or perform biased cost-aware search, all of which exacerbate identifying the best-performing model when a total cost budget is specified. This paper proposes a novel approach referred to as Budget-Aware Planning for Iterative Learners (BAPI) to solve HPO problems under a constrained cost budget. BAPI is an efficient non-myopic Bayesian optimization solution that accounts for the budget and leverages the prior knowledge about the objective function and cost function to select better configurations and to take more informed decisions during the evaluation (training). Experiments on diverse HPO benchmarks for iterative learners show that BAPI performs better than state-of-the-art baselines in most of the cases.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge