Kenney Ng
Multi-view biomedical foundation models for molecule-target and property prediction
Oct 25, 2024


Abstract:Foundation models applied to bio-molecular space hold promise to accelerate drug discovery. Molecular representation is key to building such models. Previous works have typically focused on a single representation or view of the molecules. Here, we develop a multi-view foundation model approach, that integrates molecular views of graph, image and text. Single-view foundation models are each pre-trained on a dataset of up to 200M molecules and then aggregated into combined representations. Our multi-view model is validated on a diverse set of 18 tasks, encompassing ligand-protein binding, molecular solubility, metabolism and toxicity. We show that the multi-view models perform robustly and are able to balance the strengths and weaknesses of specific views. We then apply this model to screen compounds against a large (>100 targets) set of G Protein-Coupled receptors (GPCRs). From this library of targets, we identify 33 that are related to Alzheimer's disease. On this subset, we employ our model to identify strong binders, which are validated through structure-based modeling and identification of key binding motifs.
Estimating heterogeneous treatment effect from survival outcomes via (orthogonal) censoring unbiased learning
Jan 20, 2024Abstract:Methods for estimating heterogeneous treatment effects (HTE) from observational data have largely focused on continuous or binary outcomes, with less attention paid to survival outcomes and almost none to settings with competing risks. In this work, we develop censoring unbiased transformations (CUTs) for survival outcomes both with and without competing risks.After converting time-to-event outcomes using these CUTs, direct application of HTE learners for continuous outcomes yields consistent estimates of heterogeneous cumulative incidence effects, total effects, and separable direct effects. Our CUTs enable application of a much larger set of state of the art HTE learners for censored outcomes than had previously been available, especially in competing risks settings. We provide generic model-free learner-specific oracle inequalities bounding the finite-sample excess risk. The oracle efficiency results depend on the oracle selector and estimated nuisance functions from all steps involved in the transformation. We demonstrate the empirical performance of the proposed methods in simulation studies.
Latent Space Explorer: Visual Analytics for Multimodal Latent Space Exploration
Dec 01, 2023Abstract:Machine learning models built on training data with multiple modalities can reveal new insights that are not accessible through unimodal datasets. For example, cardiac magnetic resonance images (MRIs) and electrocardiograms (ECGs) are both known to capture useful information about subjects' cardiovascular health status. A multimodal machine learning model trained from large datasets can potentially predict the onset of heart-related diseases and provide novel medical insights about the cardiovascular system. Despite the potential benefits, it is difficult for medical experts to explore multimodal representation models without visual aids and to test the predictive performance of the models on various subpopulations. To address the challenges, we developed a visual analytics system called Latent Space Explorer. Latent Space Explorer provides interactive visualizations that enable users to explore the multimodal representation of subjects, define subgroups of interest, interactively decode data with different modalities with the selected subjects, and inspect the accuracy of the embedding in downstream prediction tasks. A user study was conducted with medical experts and their feedback provided useful insights into how Latent Space Explorer can help their analysis and possible new direction for further development in the medical domain.
Efficient estimation of weighted cumulative treatment effects by double/debiased machine learning
May 03, 2023Abstract:In empirical studies with time-to-event outcomes, investigators often leverage observational data to conduct causal inference on the effect of exposure when randomized controlled trial data is unavailable. Model misspecification and lack of overlap are common issues in observational studies, and they often lead to inconsistent and inefficient estimators of the average treatment effect. Estimators targeting overlap weighted effects have been proposed to address the challenge of poor overlap, and methods enabling flexible machine learning for nuisance models address model misspecification. However, the approaches that allow machine learning for nuisance models have not been extended to the setting of weighted average treatment effects for time-to-event outcomes when there is poor overlap. In this work, we propose a class of one-step cross-fitted double/debiased machine learning estimators for the weighted cumulative causal effect as a function of restriction time. We prove that the proposed estimators are consistent, asymptotically linear, and reach semiparametric efficiency bounds under regularity conditions. Our simulations show that the proposed estimators using nonparametric machine learning nuisance models perform as well as established methods that require correctly-specified parametric nuisance models, illustrating that our estimators mitigate the need for oracle parametric nuisance models. We apply the proposed methods to real-world observational data from a UK primary care database to compare the effects of anti-diabetic drugs on cancer clinical outcomes.
Post-hoc loss-calibration for Bayesian neural networks
Jun 13, 2021



Abstract:Bayesian decision theory provides an elegant framework for acting optimally under uncertainty when tractable posterior distributions are available. Modern Bayesian models, however, typically involve intractable posteriors that are approximated with, potentially crude, surrogates. This difficulty has engendered loss-calibrated techniques that aim to learn posterior approximations that favor high-utility decisions. In this paper, focusing on Bayesian neural networks, we develop methods for correcting approximate posterior predictive distributions encouraging them to prefer high-utility decisions. In contrast to previous work, our approach is agnostic to the choice of the approximate inference algorithm, allows for efficient test time decision making through amortization, and empirically produces higher quality decisions. We demonstrate the effectiveness of our approach through controlled experiments spanning a diversity of tasks and datasets.
Blending Knowledge in Deep Recurrent Networks for Adverse Event Prediction at Hospital Discharge
Apr 09, 2021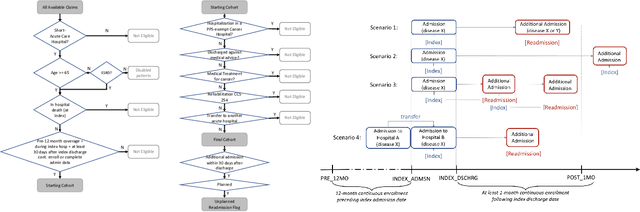
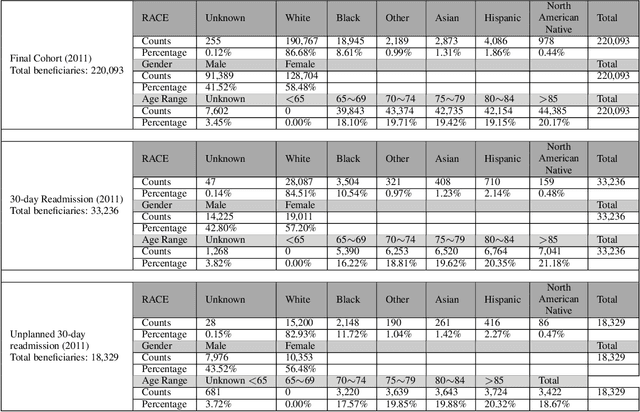
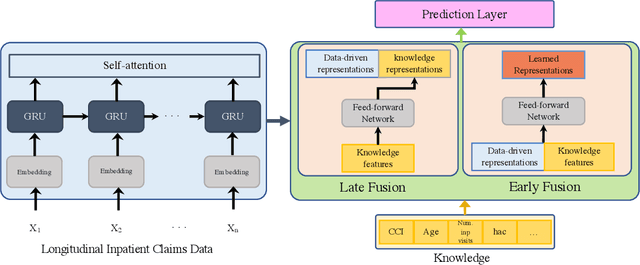

Abstract:Deep learning architectures have an extremely high-capacity for modeling complex data in a wide variety of domains. However, these architectures have been limited in their ability to support complex prediction problems using insurance claims data, such as readmission at 30 days, mainly due to data sparsity issue. Consequently, classical machine learning methods, especially those that embed domain knowledge in handcrafted features, are often on par with, and sometimes outperform, deep learning approaches. In this paper, we illustrate how the potential of deep learning can be achieved by blending domain knowledge within deep learning architectures to predict adverse events at hospital discharge, including readmissions. More specifically, we introduce a learning architecture that fuses a representation of patient data computed by a self-attention based recurrent neural network, with clinically relevant features. We conduct extensive experiments on a large claims dataset and show that the blended method outperforms the standard machine learning approaches.
Modeling Disease Progression Trajectories from Longitudinal Observational Data
Dec 09, 2020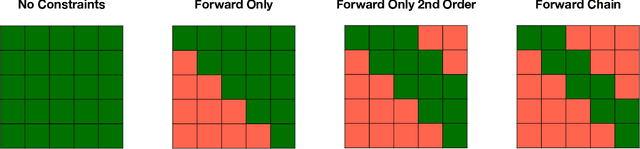

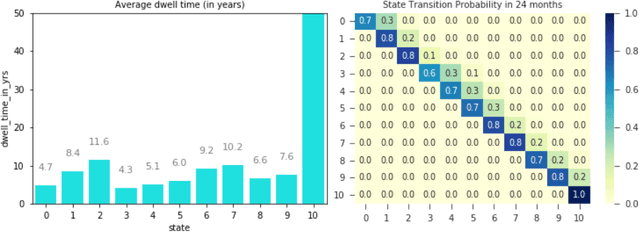
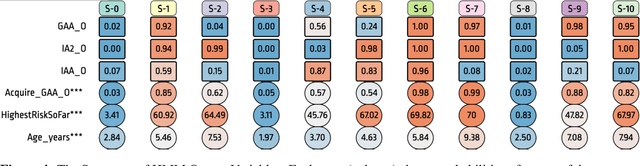
Abstract:Analyzing disease progression patterns can provide useful insights into the disease processes of many chronic conditions. These analyses may help inform recruitment for prevention trials or the development and personalization of treatments for those affected. We learn disease progression patterns using Hidden Markov Models (HMM) and distill them into distinct trajectories using visualization methods. We apply it to the domain of Type 1 Diabetes (T1D) using large longitudinal observational data from the T1DI study group. Our method discovers distinct disease progression trajectories that corroborate with recently published findings. In this paper, we describe the iterative process of developing the model. These methods may also be applied to other chronic conditions that evolve over time.
Dynamic Knowledge Distillation for Black-box Hypothesis Transfer Learning
Aug 07, 2020



Abstract:In real world applications like healthcare, it is usually difficult to build a machine learning prediction model that works universally well across different institutions. At the same time, the available model is often proprietary, i.e., neither the model parameter nor the data set used for model training is accessible. In consequence, leveraging the knowledge hidden in the available model (aka. the hypothesis) and adapting it to a local data set becomes extremely challenging. Motivated by this situation, in this paper we aim to address such a specific case within the hypothesis transfer learning framework, in which 1) the source hypothesis is a black-box model and 2) the source domain data is unavailable. In particular, we introduce a novel algorithm called dynamic knowledge distillation for hypothesis transfer learning (dkdHTL). In this method, we use knowledge distillation with instance-wise weighting mechanism to adaptively transfer the "dark" knowledge from the source hypothesis to the target domain.The weighting coefficients of the distillation loss and the standard loss are determined by the consistency between the predicted probability of the source hypothesis and the target ground-truth label.Empirical results on both transfer learning benchmark datasets and a healthcare dataset demonstrate the effectiveness of our method.
DPVis: Visual Exploration of Disease Progression Pathways
Apr 26, 2019Abstract:Clinical researchers use disease progression modeling algorithms to predict future patient status and characterize progression patterns. One approach for disease progression modeling is to describe patient status using a small number of states that represent distinctive distributions over a set of observed measures. Hidden Markov models (HMMs) and its variants are a class of models that both discover these states and make predictions concerning future states for new patients. HMMs can be trained using longitudinal observations of subjects from large-scale cohort studies, clinical trials, and electronic health records. Despite the advantages of using the algorithms for discovering interesting patterns, it still remains challenging for medical experts to interpret model outputs, complex modeling parameters, and clinically make sense of the patterns. To tackle this problem, we conducted a design study with physician scientists, statisticians, and visualization experts, with the goal to investigate disease progression pathways of certain chronic diseases, namely type 1 diabetes (T1D), Huntington's disease, Parkinson's disease, and chronic obstructive pulmonary disease (COPD). As a result, we introduce DPVis which seamlessly integrates model parameters and outcomes of HMMs into interpretable, and interactive visualizations. In this study, we demonstrate that DPVis is successful in evaluating disease progression models, visually summarizing disease states, interactively exploring disease progression patterns, and designing and comparing clinically relevant subgroup cohorts by introducing a case study on observation data from clinical studies of T1D.
Unsupervised learning with contrastive latent variable models
Nov 14, 2018



Abstract:In unsupervised learning, dimensionality reduction is an important tool for data exploration and visualization. Because these aims are typically open-ended, it can be useful to frame the problem as looking for patterns that are enriched in one dataset relative to another. These pairs of datasets occur commonly, for instance a population of interest vs. control or signal vs. signal free recordings.However, there are few methods that work on sets of data as opposed to data points or sequences. Here, we present a probabilistic model for dimensionality reduction to discover signal that is enriched in the target dataset relative to the background dataset. The data in these sets do not need to be paired or grouped beyond set membership. By using a probabilistic model where some structure is shared amongst the two datasets and some is unique to the target dataset, we are able to recover interesting structure in the latent space of the target dataset. The method also has the advantages of a probabilistic model, namely that it allows for the incorporation of prior information, handles missing data, and can be generalized to different distributional assumptions. We describe several possible variations of the model and demonstrate the application of the technique to de-noising, feature selection, and subgroup discovery settings.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge