Bum Chul Kwon
Multi-view biomedical foundation models for molecule-target and property prediction
Oct 25, 2024


Abstract:Foundation models applied to bio-molecular space hold promise to accelerate drug discovery. Molecular representation is key to building such models. Previous works have typically focused on a single representation or view of the molecules. Here, we develop a multi-view foundation model approach, that integrates molecular views of graph, image and text. Single-view foundation models are each pre-trained on a dataset of up to 200M molecules and then aggregated into combined representations. Our multi-view model is validated on a diverse set of 18 tasks, encompassing ligand-protein binding, molecular solubility, metabolism and toxicity. We show that the multi-view models perform robustly and are able to balance the strengths and weaknesses of specific views. We then apply this model to screen compounds against a large (>100 targets) set of G Protein-Coupled receptors (GPCRs). From this library of targets, we identify 33 that are related to Alzheimer's disease. On this subset, we employ our model to identify strong binders, which are validated through structure-based modeling and identification of key binding motifs.
MiMICRI: Towards Domain-centered Counterfactual Explanations of Cardiovascular Image Classification Models
Apr 24, 2024



Abstract:The recent prevalence of publicly accessible, large medical imaging datasets has led to a proliferation of artificial intelligence (AI) models for cardiovascular image classification and analysis. At the same time, the potentially significant impacts of these models have motivated the development of a range of explainable AI (XAI) methods that aim to explain model predictions given certain image inputs. However, many of these methods are not developed or evaluated with domain experts, and explanations are not contextualized in terms of medical expertise or domain knowledge. In this paper, we propose a novel framework and python library, MiMICRI, that provides domain-centered counterfactual explanations of cardiovascular image classification models. MiMICRI helps users interactively select and replace segments of medical images that correspond to morphological structures. From the counterfactuals generated, users can then assess the influence of each segment on model predictions, and validate the model against known medical facts. We evaluate this library with two medical experts. Our evaluation demonstrates that a domain-centered XAI approach can enhance the interpretability of model explanations, and help experts reason about models in terms of relevant domain knowledge. However, concerns were also surfaced about the clinical plausibility of the counterfactuals generated. We conclude with a discussion on the generalizability and trustworthiness of the MiMICRI framework, as well as the implications of our findings on the development of domain-centered XAI methods for model interpretability in healthcare contexts.
ASAP: Interpretable Analysis and Summarization of AI-generated Image Patterns at Scale
Apr 03, 2024



Abstract:Generative image models have emerged as a promising technology to produce realistic images. Despite potential benefits, concerns grow about its misuse, particularly in generating deceptive images that could raise significant ethical, legal, and societal issues. Consequently, there is growing demand to empower users to effectively discern and comprehend patterns of AI-generated images. To this end, we developed ASAP, an interactive visualization system that automatically extracts distinct patterns of AI-generated images and allows users to interactively explore them via various views. To uncover fake patterns, ASAP introduces a novel image encoder, adapted from CLIP, which transforms images into compact "distilled" representations, enriched with information for differentiating authentic and fake images. These representations generate gradients that propagate back to the attention maps of CLIP's transformer block. This process quantifies the relative importance of each pixel to image authenticity or fakeness, exposing key deceptive patterns. ASAP enables the at scale interactive analysis of these patterns through multiple, coordinated visualizations. This includes a representation overview with innovative cell glyphs to aid in the exploration and qualitative evaluation of fake patterns across a vast array of images, as well as a pattern view that displays authenticity-indicating patterns in images and quantifies their impact. ASAP supports the analysis of cutting-edge generative models with the latest architectures, including GAN-based models like proGAN and diffusion models like the latent diffusion model. We demonstrate ASAP's usefulness through two usage scenarios using multiple fake image detection benchmark datasets, revealing its ability to identify and understand hidden patterns in AI-generated images, especially in detecting fake human faces produced by diffusion-based techniques.
Latent Space Explorer: Visual Analytics for Multimodal Latent Space Exploration
Dec 01, 2023Abstract:Machine learning models built on training data with multiple modalities can reveal new insights that are not accessible through unimodal datasets. For example, cardiac magnetic resonance images (MRIs) and electrocardiograms (ECGs) are both known to capture useful information about subjects' cardiovascular health status. A multimodal machine learning model trained from large datasets can potentially predict the onset of heart-related diseases and provide novel medical insights about the cardiovascular system. Despite the potential benefits, it is difficult for medical experts to explore multimodal representation models without visual aids and to test the predictive performance of the models on various subpopulations. To address the challenges, we developed a visual analytics system called Latent Space Explorer. Latent Space Explorer provides interactive visualizations that enable users to explore the multimodal representation of subjects, define subgroups of interest, interactively decode data with different modalities with the selected subjects, and inspect the accuracy of the embedding in downstream prediction tasks. A user study was conducted with medical experts and their feedback provided useful insights into how Latent Space Explorer can help their analysis and possible new direction for further development in the medical domain.
Finspector: A Human-Centered Visual Inspection Tool for Exploring and Comparing Biases among Foundation Models
May 26, 2023Abstract:Pre-trained transformer-based language models are becoming increasingly popular due to their exceptional performance on various benchmarks. However, concerns persist regarding the presence of hidden biases within these models, which can lead to discriminatory outcomes and reinforce harmful stereotypes. To address this issue, we propose Finspector, a human-centered visual inspection tool designed to detect biases in different categories through log-likelihood scores generated by language models. The goal of the tool is to enable researchers to easily identify potential biases using visual analytics, ultimately contributing to a fairer and more just deployment of these models in both academic and industrial settings. Finspector is available at https://github.com/IBM/finspector.
DASH: Visual Analytics for Debiasing Image Classification via User-Driven Synthetic Data Augmentation
Sep 14, 2022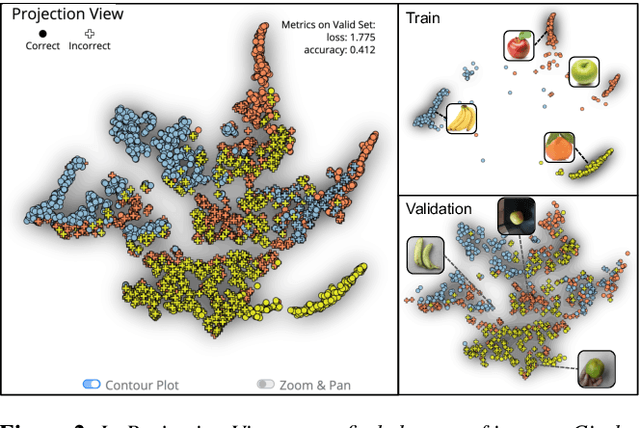
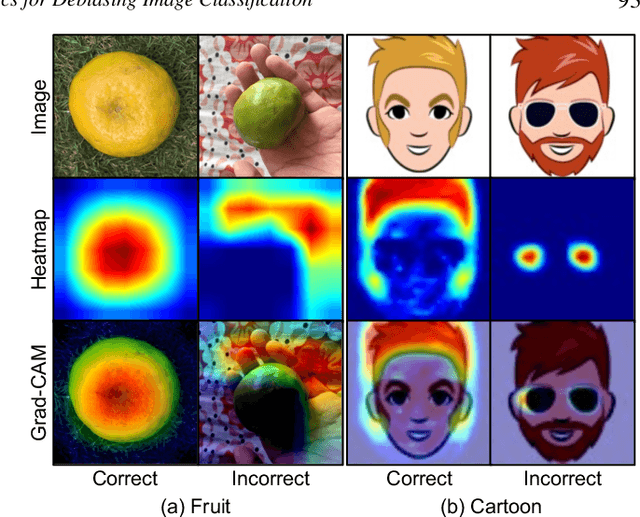
Abstract:Image classification models often learn to predict a class based on irrelevant co-occurrences between input features and an output class in training data. We call the unwanted correlations "data biases," and the visual features causing data biases "bias factors." It is challenging to identify and mitigate biases automatically without human intervention. Therefore, we conducted a design study to find a human-in-the-loop solution. First, we identified user tasks that capture the bias mitigation process for image classification models with three experts. Then, to support the tasks, we developed a visual analytics system called DASH that allows users to visually identify bias factors, to iteratively generate synthetic images using a state-of-the-art image-to-image translation model, and to supervise the model training process for improving the classification accuracy. Our quantitative evaluation and qualitative study with ten participants demonstrate the usefulness of DASH and provide lessons for future work.
Modeling Disease Progression Trajectories from Longitudinal Observational Data
Dec 09, 2020

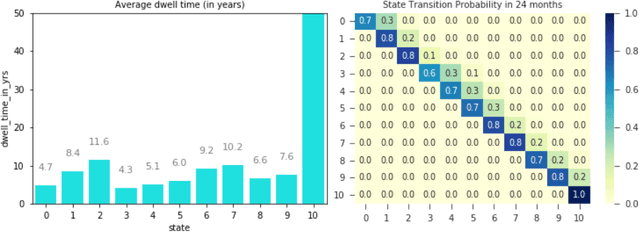
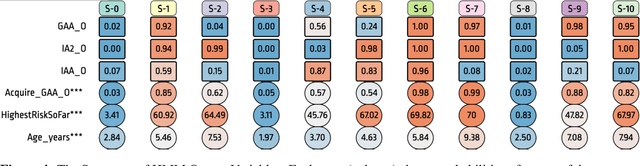
Abstract:Analyzing disease progression patterns can provide useful insights into the disease processes of many chronic conditions. These analyses may help inform recruitment for prevention trials or the development and personalization of treatments for those affected. We learn disease progression patterns using Hidden Markov Models (HMM) and distill them into distinct trajectories using visualization methods. We apply it to the domain of Type 1 Diabetes (T1D) using large longitudinal observational data from the T1DI study group. Our method discovers distinct disease progression trajectories that corroborate with recently published findings. In this paper, we describe the iterative process of developing the model. These methods may also be applied to other chronic conditions that evolve over time.
SANVis: Visual Analytics for Understanding Self-Attention Networks
Sep 13, 2019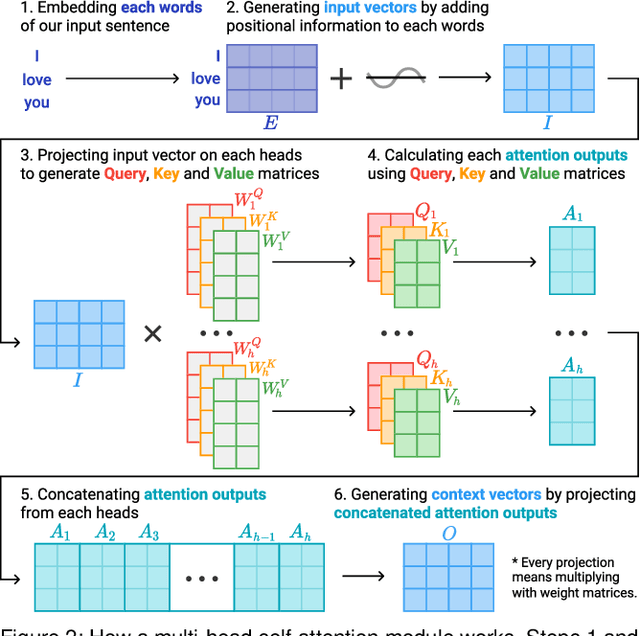
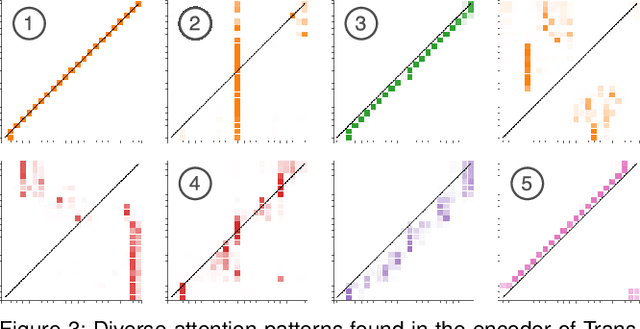
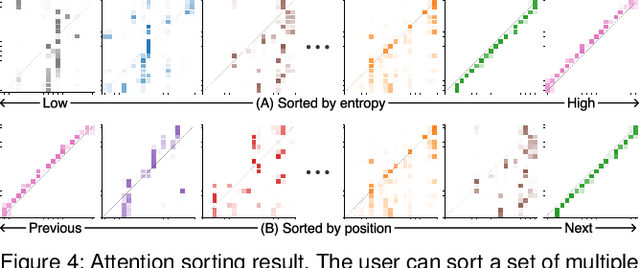
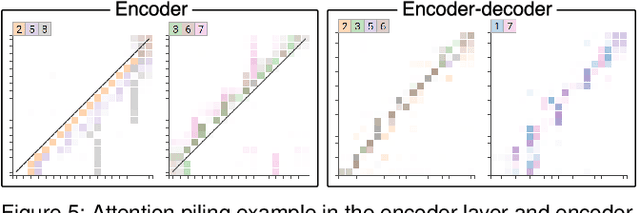
Abstract:Attention networks, a deep neural network architecture inspired by humans' attention mechanism, have seen significant success in image captioning, machine translation, and many other applications. Recently, they have been further evolved into an advanced approach called multi-head self-attention networks, which can encode a set of input vectors, e.g., word vectors in a sentence, into another set of vectors. Such encoding aims at simultaneously capturing diverse syntactic and semantic features within a set, each of which corresponds to a particular attention head, forming altogether multi-head attention. Meanwhile, the increased model complexity prevents users from easily understanding and manipulating the inner workings of models. To tackle the challenges, we present a visual analytics system called SANVis, which helps users understand the behaviors and the characteristics of multi-head self-attention networks. Using a state-of-the-art self-attention model called Transformer, we demonstrate usage scenarios of SANVis in machine translation tasks. Our system is available at http://short.sanvis.org
DPVis: Visual Exploration of Disease Progression Pathways
Apr 26, 2019Abstract:Clinical researchers use disease progression modeling algorithms to predict future patient status and characterize progression patterns. One approach for disease progression modeling is to describe patient status using a small number of states that represent distinctive distributions over a set of observed measures. Hidden Markov models (HMMs) and its variants are a class of models that both discover these states and make predictions concerning future states for new patients. HMMs can be trained using longitudinal observations of subjects from large-scale cohort studies, clinical trials, and electronic health records. Despite the advantages of using the algorithms for discovering interesting patterns, it still remains challenging for medical experts to interpret model outputs, complex modeling parameters, and clinically make sense of the patterns. To tackle this problem, we conducted a design study with physician scientists, statisticians, and visualization experts, with the goal to investigate disease progression pathways of certain chronic diseases, namely type 1 diabetes (T1D), Huntington's disease, Parkinson's disease, and chronic obstructive pulmonary disease (COPD). As a result, we introduce DPVis which seamlessly integrates model parameters and outcomes of HMMs into interpretable, and interactive visualizations. In this study, we demonstrate that DPVis is successful in evaluating disease progression models, visually summarizing disease states, interactively exploring disease progression patterns, and designing and comparing clinically relevant subgroup cohorts by introducing a case study on observation data from clinical studies of T1D.
RetainVis: Visual Analytics with Interpretable and Interactive Recurrent Neural Networks on Electronic Medical Records
Oct 23, 2018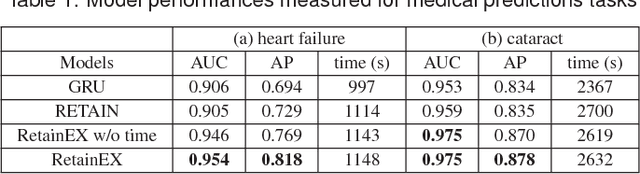
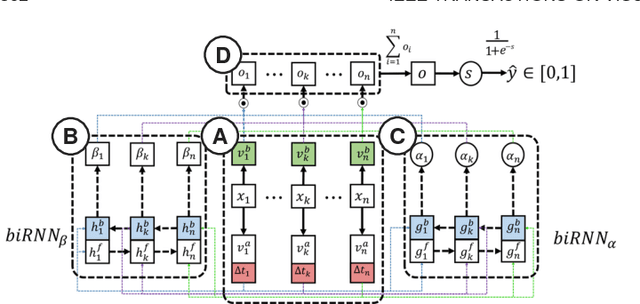
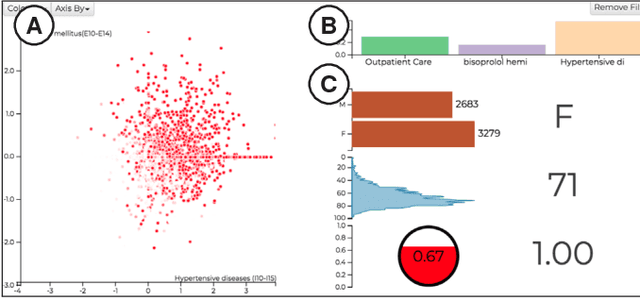
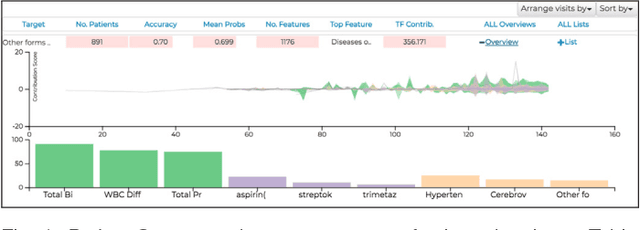
Abstract:We have recently seen many successful applications of recurrent neural networks (RNNs) on electronic medical records (EMRs), which contain histories of patients' diagnoses, medications, and other various events, in order to predict the current and future states of patients. Despite the strong performance of RNNs, it is often challenging for users to understand why the model makes a particular prediction. Such black-box nature of RNNs can impede its wide adoption in clinical practice. Furthermore, we have no established methods to interactively leverage users' domain expertise and prior knowledge as inputs for steering the model. Therefore, our design study aims to provide a visual analytics solution to increase interpretability and interactivity of RNNs via a joint effort of medical experts, artificial intelligence scientists, and visual analytics researchers. Following the iterative design process between the experts, we design, implement, and evaluate a visual analytics tool called RetainVis, which couples a newly improved, interpretable and interactive RNN-based model called RetainEX and visualizations for users' exploration of EMR data in the context of prediction tasks. Our study shows the effective use of RetainVis for gaining insights into how individual medical codes contribute to making risk predictions, using EMRs of patients with heart failure and cataract symptoms. Our study also demonstrates how we made substantial changes to the state-of-the-art RNN model called RETAIN in order to make use of temporal information and increase interactivity. This study will provide a useful guideline for researchers that aim to design an interpretable and interactive visual analytics tool for RNNs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge