Jaehyo Yoo
Format Inertia: A Failure Mechanism of LLMs in Medical Pre-Consultation
Oct 02, 2025Abstract:Recent advances in Large Language Models (LLMs) have brought significant improvements to various service domains, including chatbots and medical pre-consultation applications. In the healthcare domain, the most common approach for adapting LLMs to multi-turn dialogue generation is Supervised Fine-Tuning (SFT). However, datasets for SFT in tasks like medical pre-consultation typically exhibit a skewed turn-count distribution. Training on such data induces a novel failure mechanism we term **Format Inertia**, where models tend to generate repetitive, format-correct, but diagnostically uninformative questions in long medical dialogues. To mitigate this observed failure mechanism, we adopt a simple, data-centric method that rebalances the turn-count distribution of the training dataset. Experimental results show that our approach substantially alleviates Format Inertia in medical pre-consultation.
Taxonomy of Comprehensive Safety for Clinical Agents
Sep 26, 2025Abstract:Safety is a paramount concern in clinical chatbot applications, where inaccurate or harmful responses can lead to serious consequences. Existing methods--such as guardrails and tool calling--often fall short in addressing the nuanced demands of the clinical domain. In this paper, we introduce TACOS (TAxonomy of COmprehensive Safety for Clinical Agents), a fine-grained, 21-class taxonomy that integrates safety filtering and tool selection into a single user intent classification step. TACOS is a taxonomy that can cover a wide spectrum of clinical and non-clinical queries, explicitly modeling varying safety thresholds and external tool dependencies. To validate our framework, we curate a TACOS-annotated dataset and perform extensive experiments. Our results demonstrate the value of a new taxonomy specialized for clinical agent settings, and reveal useful insights about train data distribution and pretrained knowledge of base models.
Automatic Creation of Named Entity Recognition Datasets by Querying Phrase Representations
Oct 14, 2022



Abstract:Most weakly supervised named entity recognition (NER) models rely on domain-specific dictionaries provided by experts. This approach is infeasible in many domains where dictionaries do not exist. While a phrase retrieval model was used to construct pseudo-dictionaries with entities retrieved from Wikipedia automatically in a recent study, these dictionaries often have limited coverage because the retriever is likely to retrieve popular entities rather than rare ones. In this study, a phrase embedding search to efficiently create high-coverage dictionaries is presented. Specifically, the reformulation of natural language queries into phrase representations allows the retriever to search a space densely populated with various entities. In addition, we present a novel framework, HighGEN, that generates NER datasets with high-coverage dictionaries obtained using the phrase embedding search. HighGEN generates weak labels based on the distance between the embeddings of a candidate phrase and target entity type to reduce the noise in high-coverage dictionaries. We compare HighGEN with current weakly supervised NER models on six NER benchmarks and demonstrate the superiority of our models.
Lack of Fluency is Hurting Your Translation Model
May 24, 2022
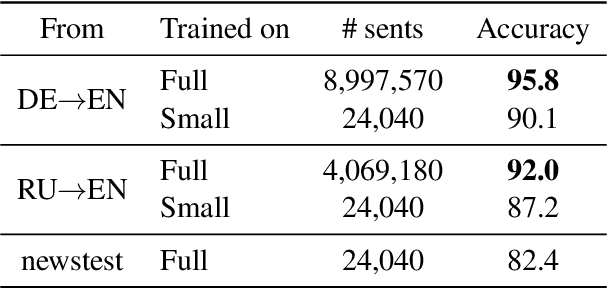
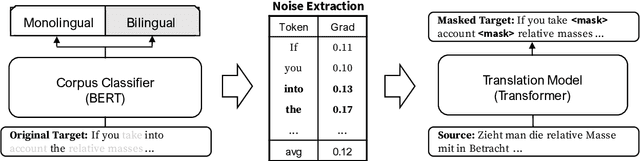
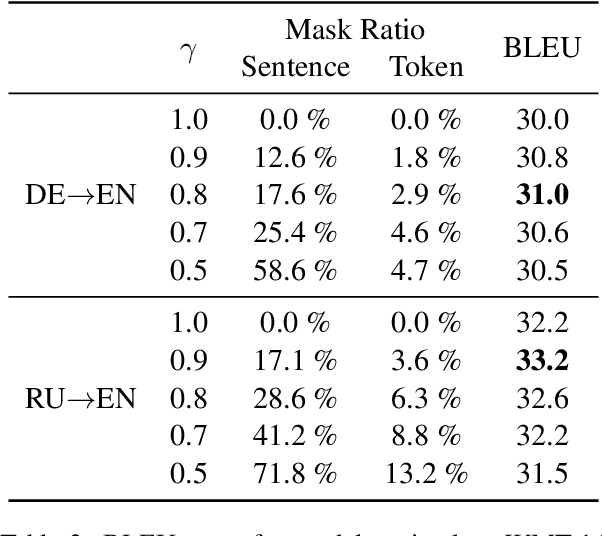
Abstract:Many machine translation models are trained on bilingual corpus, which consist of aligned sentence pairs from two different languages with same semantic. However, there is a qualitative discrepancy between train and test set in bilingual corpus. While the most train sentences are created via automatic techniques such as crawling and sentence-alignment methods, the test sentences are annotated with the consideration of fluency by human. We suppose this discrepancy in training corpus will yield performance drop of translation model. In this work, we define \textit{fluency noise} to determine which parts of train sentences cause them to seem unnatural. We show that \textit{fluency noise} can be detected by simple gradient-based method with pre-trained classifier. By removing \textit{fluency noise} in train sentences, our final model outperforms the baseline on WMT-14 DE$\rightarrow$EN and RU$\rightarrow$EN. We also show the compatibility with back-translation augmentation, which has been commonly used to improve the fluency of the translation model. At last, the qualitative analysis of \textit{fluency noise} provides the insight of what points we should focus on.
Simple Questions Generate Named Entity Recognition Datasets
Dec 16, 2021



Abstract:Named entity recognition (NER) is a task of extracting named entities of specific types from text. Current NER models often rely on human-annotated datasets requiring the vast engagement of professional knowledge on the target domain and entities. This work introduces an ask-to-generate approach, which automatically generates NER datasets by asking simple natural language questions that reflect the needs for entity types (e.g., Which disease?) to an open-domain question answering system. Without using any in-domain resources (i.e., training sentences, labels, or in-domain dictionaries), our models solely trained on our generated datasets largely outperform previous weakly supervised models on six NER benchmarks across four different domains. Surprisingly, on NCBI-disease, our model achieves 75.5 F1 score and even outperforms the previous best weakly supervised model by 4.1 F1 score, which utilizes a rich in-domain dictionary provided by domain experts. Formulating the needs of NER with natural language also allows us to build NER models for fine-grained entity types such as Award, where our model even outperforms fully supervised models. On three few-shot NER benchmarks, our model achieves new state-of-the-art performance.
Transferability of Natural Language Inference to Biomedical Question Answering
Jul 01, 2020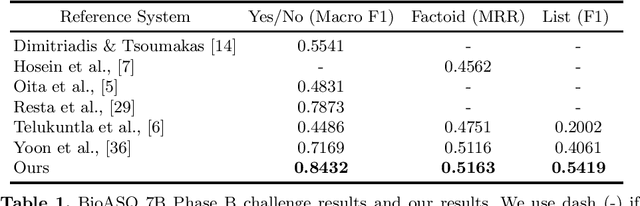
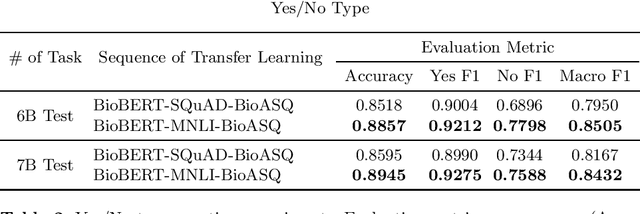
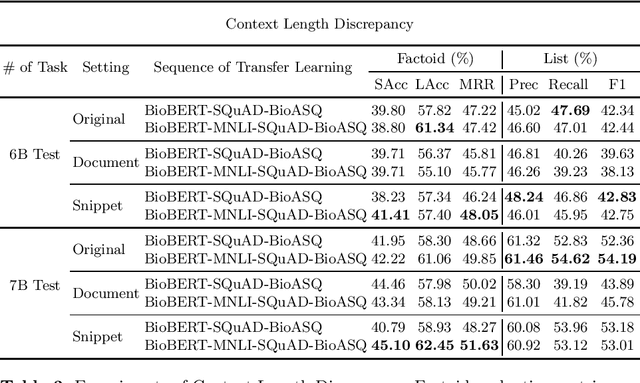
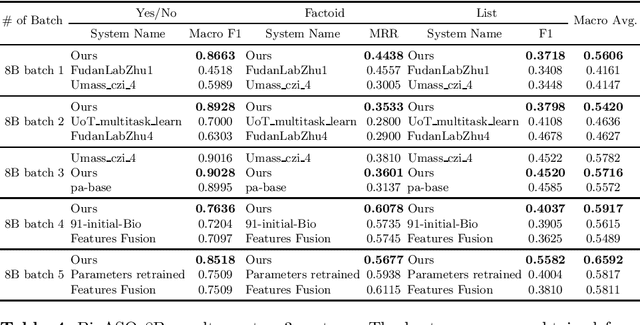
Abstract:Biomedical question answering (QA) is a challenging problem due to the scarcity of data and the requirement of domain expertise. Growing interests of using pre-trained language models with transfer learning address the issue to some extent. Recently, learning linguistic knowledge of entailment in sentence pairs enhances the performance in general domain QA by leveraging such transferability between the two tasks. In this paper, we focus on facilitating the transferability by unifying the experimental setup from natural language inference (NLI) to biomedical QA. We observe that transferring from entailment data shows effective performance on Yes/No (+5.59%), Factoid (+0.53%), List (+13.58%) type questions compared to previous challenge reports (BioASQ 7B Phase B). We also observe that our method generally performs well in the 8th BioASQ Challenge (Phase B). For sequential transfer learning, the order of how tasks are fine-tuned is important. In factoid- and list-type questions, we thoroughly analyze an intrinsic limitation of the extractive QA setting when these questions are converted to the same format of the Stanford Question Answering Dataset (SQuAD).
SANVis: Visual Analytics for Understanding Self-Attention Networks
Sep 13, 2019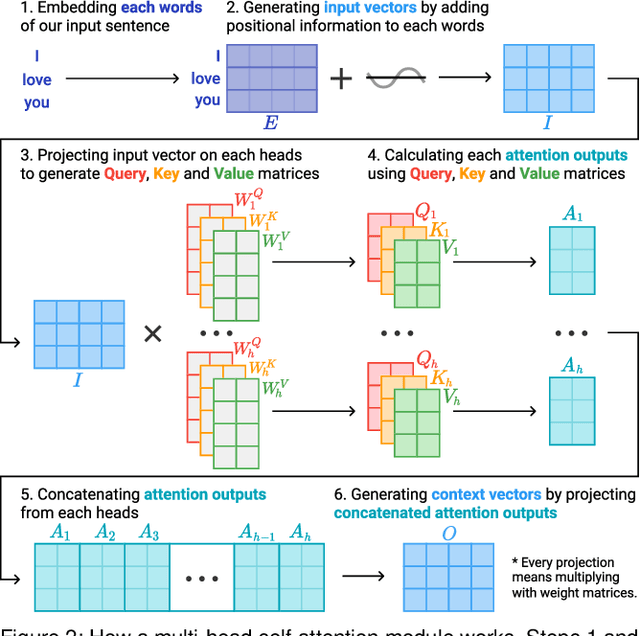
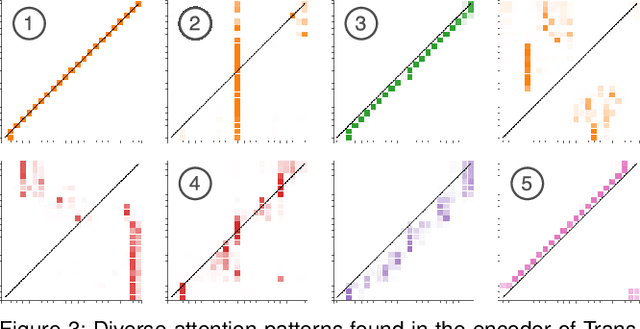
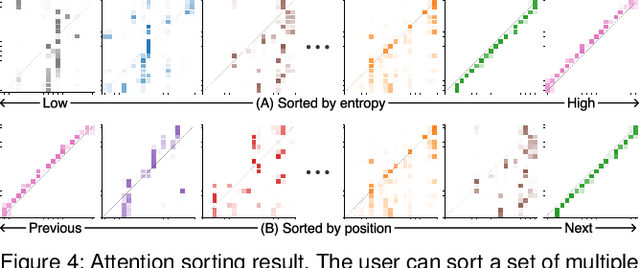
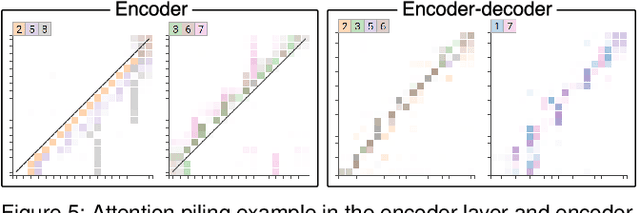
Abstract:Attention networks, a deep neural network architecture inspired by humans' attention mechanism, have seen significant success in image captioning, machine translation, and many other applications. Recently, they have been further evolved into an advanced approach called multi-head self-attention networks, which can encode a set of input vectors, e.g., word vectors in a sentence, into another set of vectors. Such encoding aims at simultaneously capturing diverse syntactic and semantic features within a set, each of which corresponds to a particular attention head, forming altogether multi-head attention. Meanwhile, the increased model complexity prevents users from easily understanding and manipulating the inner workings of models. To tackle the challenges, we present a visual analytics system called SANVis, which helps users understand the behaviors and the characteristics of multi-head self-attention networks. Using a state-of-the-art self-attention model called Transformer, we demonstrate usage scenarios of SANVis in machine translation tasks. Our system is available at http://short.sanvis.org
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge