Mujeen Sung
Trustworthy Agents for Electronic Health Records through Confidence Estimation
Aug 26, 2025


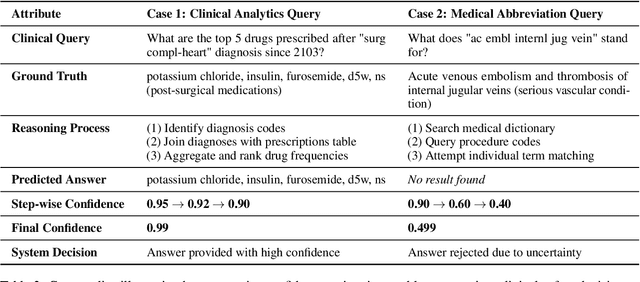
Abstract:Large language models (LLMs) show promise for extracting information from Electronic Health Records (EHR) and supporting clinical decisions. However, deployment in clinical settings faces challenges due to hallucination risks. We propose Hallucination Controlled Accuracy at k% (HCAcc@k%), a novel metric quantifying the accuracy-reliability trade-off at varying confidence thresholds. We introduce TrustEHRAgent, a confidence-aware agent incorporating stepwise confidence estimation for clinical question answering. Experiments on MIMIC-III and eICU datasets show TrustEHRAgent outperforms baselines under strict reliability constraints, achieving improvements of 44.23%p and 25.34%p at HCAcc@70% while baseline methods fail at these thresholds. These results highlight limitations of traditional accuracy metrics in evaluating healthcare AI agents. Our work contributes to developing trustworthy clinical agents that deliver accurate information or transparently express uncertainty when confidence is low.
KU AIGEN ICL EDI@BC8 Track 3: Advancing Phenotype Named Entity Recognition and Normalization for Dysmorphology Physical Examination Reports
Jan 16, 2025

Abstract:The objective of BioCreative8 Track 3 is to extract phenotypic key medical findings embedded within EHR texts and subsequently normalize these findings to their Human Phenotype Ontology (HPO) terms. However, the presence of diverse surface forms in phenotypic findings makes it challenging to accurately normalize them to the correct HPO terms. To address this challenge, we explored various models for named entity recognition and implemented data augmentation techniques such as synonym marginalization to enhance the normalization step. Our pipeline resulted in an exact extraction and normalization F1 score 2.6\% higher than the mean score of all submissions received in response to the challenge. Furthermore, in terms of the normalization F1 score, our approach surpassed the average performance by 1.9\%. These findings contribute to the advancement of automated medical data extraction and normalization techniques, showcasing potential pathways for future research and application in the biomedical domain.
Rationale-Guided Retrieval Augmented Generation for Medical Question Answering
Nov 01, 2024



Abstract:Large language models (LLM) hold significant potential for applications in biomedicine, but they struggle with hallucinations and outdated knowledge. While retrieval-augmented generation (RAG) is generally employed to address these issues, it also has its own set of challenges: (1) LLMs are vulnerable to irrelevant or incorrect context, (2) medical queries are often not well-targeted for helpful information, and (3) retrievers are prone to bias toward the specific source corpus they were trained on. In this study, we present RAG$^2$ (RAtionale-Guided RAG), a new framework for enhancing the reliability of RAG in biomedical contexts. RAG$^2$ incorporates three key innovations: a small filtering model trained on perplexity-based labels of rationales, which selectively augments informative snippets of documents while filtering out distractors; LLM-generated rationales as queries to improve the utility of retrieved snippets; a structure designed to retrieve snippets evenly from a comprehensive set of four biomedical corpora, effectively mitigating retriever bias. Our experiments demonstrate that RAG$^2$ improves the state-of-the-art LLMs of varying sizes, with improvements of up to 6.1\%, and it outperforms the previous best medical RAG model by up to 5.6\% across three medical question-answering benchmarks. Our code is available at https://github.com/dmis-lab/RAG2.
Structured List-Grounded Question Answering
Oct 04, 2024Abstract:Document-grounded dialogue systems aim to answer user queries by leveraging external information. Previous studies have mainly focused on handling free-form documents, often overlooking structured data such as lists, which can represent a range of nuanced semantic relations. Motivated by the observation that even advanced language models like GPT-3.5 often miss semantic cues from lists, this paper aims to enhance question answering (QA) systems for better interpretation and use of structured lists. To this end, we introduce the LIST2QA dataset, a novel benchmark to evaluate the ability of QA systems to respond effectively using list information. This dataset is created from unlabeled customer service documents using language models and model-based filtering processes to enhance data quality, and can be used to fine-tune and evaluate QA models. Apart from directly generating responses through fine-tuned models, we further explore the explicit use of Intermediate Steps for Lists (ISL), aligning list items with user backgrounds to better reflect how humans interpret list items before generating responses. Our experimental results demonstrate that models trained on LIST2QA with our ISL approach outperform baselines across various metrics. Specifically, our fine-tuned Flan-T5-XL model shows increases of 3.1% in ROUGE-L, 4.6% in correctness, 4.5% in faithfulness, and 20.6% in completeness compared to models without applying filtering and the proposed ISL method.
Learning from Negative Samples in Generative Biomedical Entity Linking
Aug 29, 2024



Abstract:Generative models have become widely used in biomedical entity linking (BioEL) due to their excellent performance and efficient memory usage. However, these models are usually trained only with positive samples--entities that match the input mention's identifier--and do not explicitly learn from hard negative samples, which are entities that look similar but have different meanings. To address this limitation, we introduce ANGEL (Learning from Negative Samples in Generative Biomedical Entity Linking), the first framework that trains generative BioEL models using negative samples. Specifically, a generative model is initially trained to generate positive samples from the knowledge base for given input entities. Subsequently, both correct and incorrect outputs are gathered from the model's top-k predictions. The model is then updated to prioritize the correct predictions through direct preference optimization. Our models fine-tuned with ANGEL outperform the previous best baseline models by up to an average top-1 accuracy of 1.4% on five benchmarks. When incorporating our framework into pre-training, the performance improvement further increases to 1.7%, demonstrating its effectiveness in both the pre-training and fine-tuning stages. Our code is available at https://github.com/dmis-lab/ANGEL.
Infusing Environmental Captions for Long-Form Video Language Grounding
Aug 06, 2024



Abstract:In this work, we tackle the problem of long-form video-language grounding (VLG). Given a long-form video and a natural language query, a model should temporally localize the precise moment that answers the query. Humans can easily solve VLG tasks, even with arbitrarily long videos, by discarding irrelevant moments using extensive and robust knowledge gained from experience. Unlike humans, existing VLG methods are prone to fall into superficial cues learned from small-scale datasets, even when they are within irrelevant frames. To overcome this challenge, we propose EI-VLG, a VLG method that leverages richer textual information provided by a Multi-modal Large Language Model (MLLM) as a proxy for human experiences, helping to effectively exclude irrelevant frames. We validate the effectiveness of the proposed method via extensive experiments on a challenging EgoNLQ benchmark.
CookingSense: A Culinary Knowledgebase with Multidisciplinary Assertions
May 01, 2024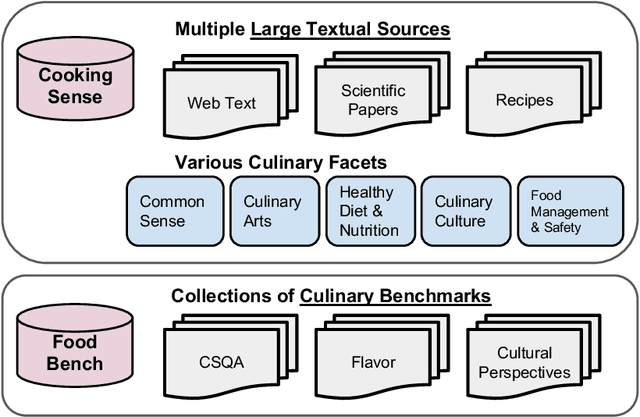
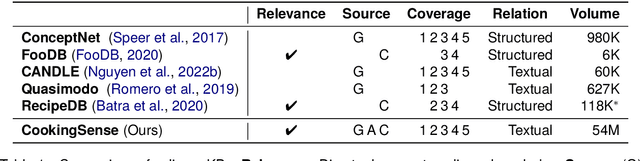
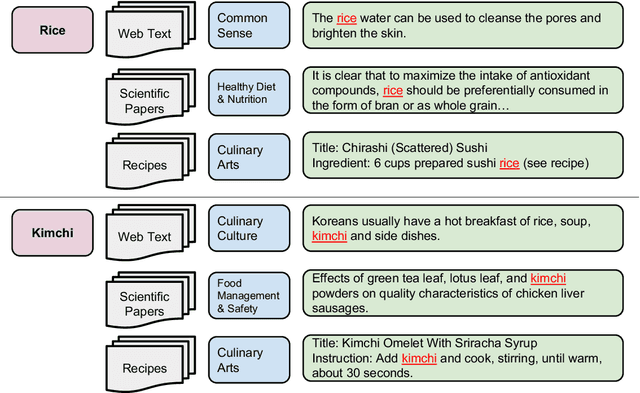
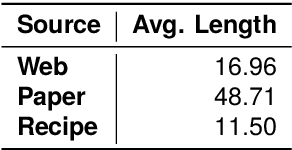
Abstract:This paper introduces CookingSense, a descriptive collection of knowledge assertions in the culinary domain extracted from various sources, including web data, scientific papers, and recipes, from which knowledge covering a broad range of aspects is acquired. CookingSense is constructed through a series of dictionary-based filtering and language model-based semantic filtering techniques, which results in a rich knowledgebase of multidisciplinary food-related assertions. Additionally, we present FoodBench, a novel benchmark to evaluate culinary decision support systems. From evaluations with FoodBench, we empirically prove that CookingSense improves the performance of retrieval augmented language models. We also validate the quality and variety of assertions in CookingSense through qualitative analysis.
Improving Medical Reasoning through Retrieval and Self-Reflection with Retrieval-Augmented Large Language Models
Jan 27, 2024



Abstract:Recent proprietary large language models (LLMs), such as GPT-4, have achieved a milestone in tackling diverse challenges in the biomedical domain, ranging from multiple-choice questions to long-form generations. To address challenges that still cannot be handled with the encoded knowledge of LLMs, various retrieval-augmented generation (RAG) methods have been developed by searching documents from the knowledge corpus and appending them unconditionally or selectively to the input of LLMs for generation. However, when applying existing methods to different domain-specific problems, poor generalization becomes apparent, leading to fetching incorrect documents or making inaccurate judgments. In this paper, we introduce Self-BioRAG, a framework reliable for biomedical text that specializes in generating explanations, retrieving domain-specific documents, and self-reflecting generated responses. We utilize 84k filtered biomedical instruction sets to train Self-BioRAG that can assess its generated explanations with customized reflective tokens. Our work proves that domain-specific components, such as a retriever, domain-related document corpus, and instruction sets are necessary for adhering to domain-related instructions. Using three major medical question-answering benchmark datasets, experimental results of Self-BioRAG demonstrate significant performance gains by achieving a 7.2% absolute improvement on average over the state-of-the-art open-foundation model with a parameter size of 7B or less. Overall, we analyze that Self-BioRAG finds the clues in the question, retrieves relevant documents if needed, and understands how to answer with information from retrieved documents and encoded knowledge as a medical expert does. We release our data and code for training our framework components and model weights (7B and 13B) to enhance capabilities in biomedical and clinical domains.
KU-DMIS-MSRA at RadSum23: Pre-trained Vision-Language Model for Radiology Report Summarization
Jul 10, 2023Abstract:In this paper, we introduce CheXOFA, a new pre-trained vision-language model (VLM) for the chest X-ray domain. Our model is initially pre-trained on various multimodal datasets within the general domain before being transferred to the chest X-ray domain. Following a prominent VLM, we unify various domain-specific tasks into a simple sequence-to-sequence schema. It enables the model to effectively learn the required knowledge and skills from limited resources in the domain. Demonstrating superior performance on the benchmark datasets provided by the BioNLP shared task, our model benefits from its training across multiple tasks and domains. With subtle techniques including ensemble and factual calibration, our system achieves first place on the RadSum23 leaderboard for the hidden test set.
Pre-training Intent-Aware Encoders for Zero- and Few-Shot Intent Classification
May 24, 2023Abstract:Intent classification (IC) plays an important role in task-oriented dialogue systems as it identifies user intents from given utterances. However, models trained on limited annotations for IC often suffer from a lack of generalization to unseen intent classes. We propose a novel pre-training method for text encoders that uses contrastive learning with intent psuedo-labels to produce embeddings that are well-suited for IC tasks. By applying this pre-training strategy, we also introduce the pre-trained intent-aware encoder (PIE). Specifically, we first train a tagger to identify key phrases within utterances that are crucial for interpreting intents. We then use these extracted phrases to create examples for pre-training a text encoder in a contrastive manner. As a result, our PIE model achieves up to 5.4% and 4.0% higher accuracy than the previous state-of-the-art pre-trained sentence encoder for the N-way zero- and one-shot settings on four IC datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge