Sunkyu Kim
KU AIGEN ICL EDI@BC8 Track 3: Advancing Phenotype Named Entity Recognition and Normalization for Dysmorphology Physical Examination Reports
Jan 16, 2025

Abstract:The objective of BioCreative8 Track 3 is to extract phenotypic key medical findings embedded within EHR texts and subsequently normalize these findings to their Human Phenotype Ontology (HPO) terms. However, the presence of diverse surface forms in phenotypic findings makes it challenging to accurately normalize them to the correct HPO terms. To address this challenge, we explored various models for named entity recognition and implemented data augmentation techniques such as synonym marginalization to enhance the normalization step. Our pipeline resulted in an exact extraction and normalization F1 score 2.6\% higher than the mean score of all submissions received in response to the challenge. Furthermore, in terms of the normalization F1 score, our approach surpassed the average performance by 1.9\%. These findings contribute to the advancement of automated medical data extraction and normalization techniques, showcasing potential pathways for future research and application in the biomedical domain.
BioBERT: a pre-trained biomedical language representation model for biomedical text mining
Feb 03, 2019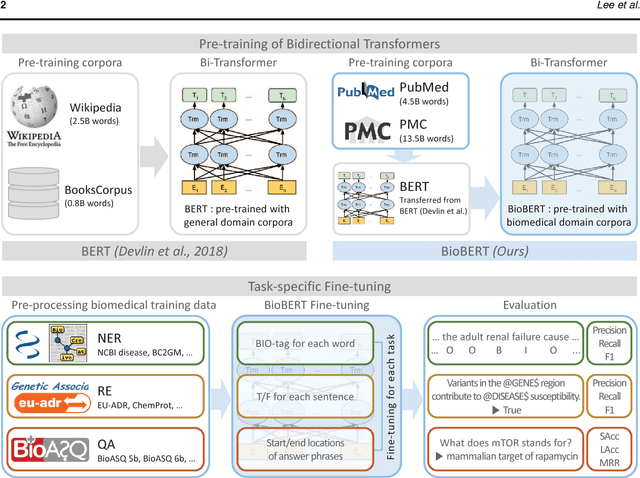
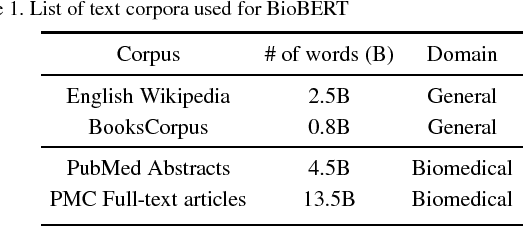

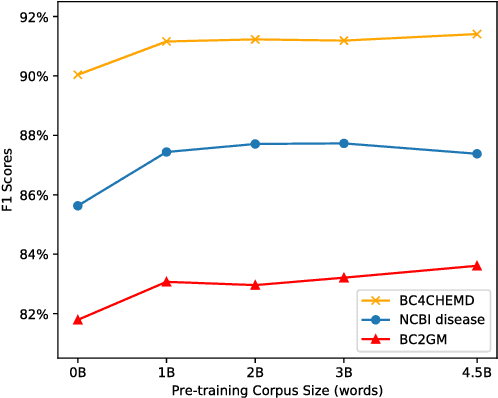
Abstract:Biomedical text mining is becoming increasingly important as the number of biomedical documents rapidly grows. With the progress in machine learning, extracting valuable information from biomedical literature has gained popularity among researchers, and deep learning has boosted the development of effective biomedical text mining models. However, as deep learning models require a large amount of training data, applying deep learning to biomedical text mining is often unsuccessful due to the lack of training data in biomedical fields. Recent researches on training contextualized language representation models on text corpora shed light on the possibility of leveraging a large number of unannotated biomedical text corpora. We introduce BioBERT (Bidirectional Encoder Representations from Transformers for Biomedical Text Mining), which is a domain specific language representation model pre-trained on large-scale biomedical corpora. Based on the BERT architecture, BioBERT effectively transfers the knowledge from a large amount of biomedical texts to biomedical text mining models with minimal task-specific architecture modifications. While BERT shows competitive performances with previous state-of-the-art models, BioBERT significantly outperforms them on the following three representative biomedical text mining tasks: biomedical named entity recognition (0.51% absolute improvement), biomedical relation extraction (3.49% absolute improvement), and biomedical question answering (9.61% absolute improvement). We make the pre-trained weights of BioBERT freely available at https://github.com/naver/biobert-pretrained, and the source code for fine-tuning BioBERT available at https://github.com/dmis-lab/biobert.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge