Chan Ho So
HATS: A Hierarchical Graph Attention Network for Stock Movement Prediction
Aug 24, 2019



Abstract:Many researchers both in academia and industry have long been interested in the stock market. Numerous approaches were developed to accurately predict future trends in stock prices. Recently, there has been a growing interest in utilizing graph-structured data in computer science research communities. Methods that use relational data for stock market prediction have been recently proposed, but they are still in their infancy. First, the quality of collected information from different types of relations can vary considerably. No existing work has focused on the effect of using different types of relations on stock market prediction or finding an effective way to selectively aggregate information on different relation types. Furthermore, existing works have focused on only individual stock prediction which is similar to the node classification task. To address this, we propose a hierarchical attention network for stock prediction (HATS) which uses relational data for stock market prediction. Our HATS method selectively aggregates information on different relation types and adds the information to the representations of each company. Specifically, node representations are initialized with features extracted from a feature extraction module. HATS is used as a relational modeling module with initialized node representations. Then, node representations with the added information are fed into a task-specific layer. Our method is used for predicting not only individual stock prices but also market index movements, which is similar to the graph classification task. The experimental results show that performance can change depending on the relational data used. HATS which can automatically select information outperformed all the existing methods.
BioBERT: a pre-trained biomedical language representation model for biomedical text mining
Feb 03, 2019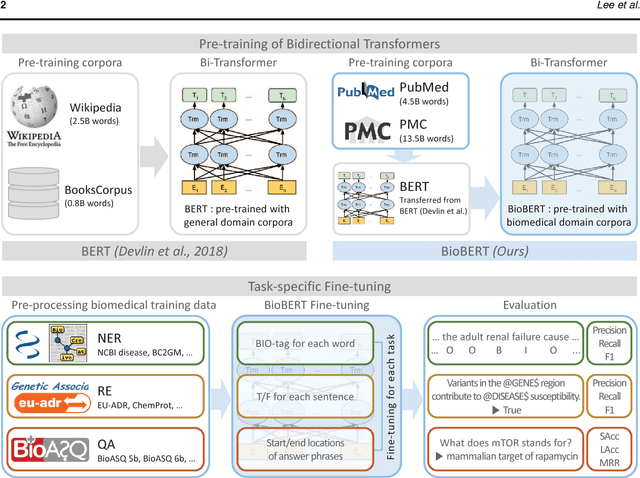
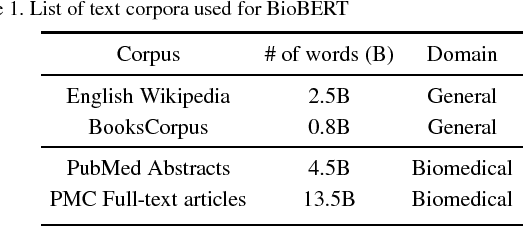

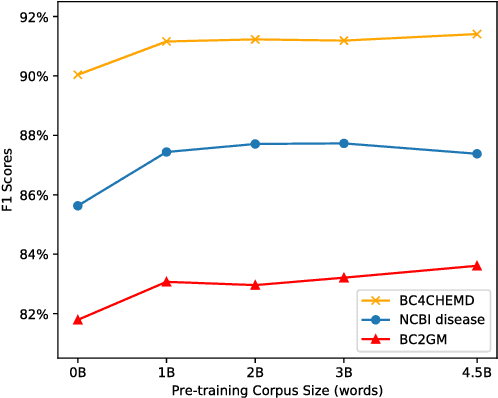
Abstract:Biomedical text mining is becoming increasingly important as the number of biomedical documents rapidly grows. With the progress in machine learning, extracting valuable information from biomedical literature has gained popularity among researchers, and deep learning has boosted the development of effective biomedical text mining models. However, as deep learning models require a large amount of training data, applying deep learning to biomedical text mining is often unsuccessful due to the lack of training data in biomedical fields. Recent researches on training contextualized language representation models on text corpora shed light on the possibility of leveraging a large number of unannotated biomedical text corpora. We introduce BioBERT (Bidirectional Encoder Representations from Transformers for Biomedical Text Mining), which is a domain specific language representation model pre-trained on large-scale biomedical corpora. Based on the BERT architecture, BioBERT effectively transfers the knowledge from a large amount of biomedical texts to biomedical text mining models with minimal task-specific architecture modifications. While BERT shows competitive performances with previous state-of-the-art models, BioBERT significantly outperforms them on the following three representative biomedical text mining tasks: biomedical named entity recognition (0.51% absolute improvement), biomedical relation extraction (3.49% absolute improvement), and biomedical question answering (9.61% absolute improvement). We make the pre-trained weights of BioBERT freely available at https://github.com/naver/biobert-pretrained, and the source code for fine-tuning BioBERT available at https://github.com/dmis-lab/biobert.
CollaboNet: collaboration of deep neural networks for biomedical named entity recognition
Sep 21, 2018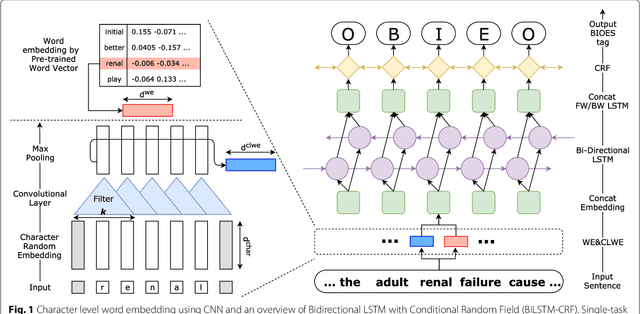
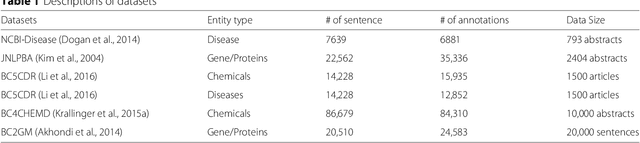
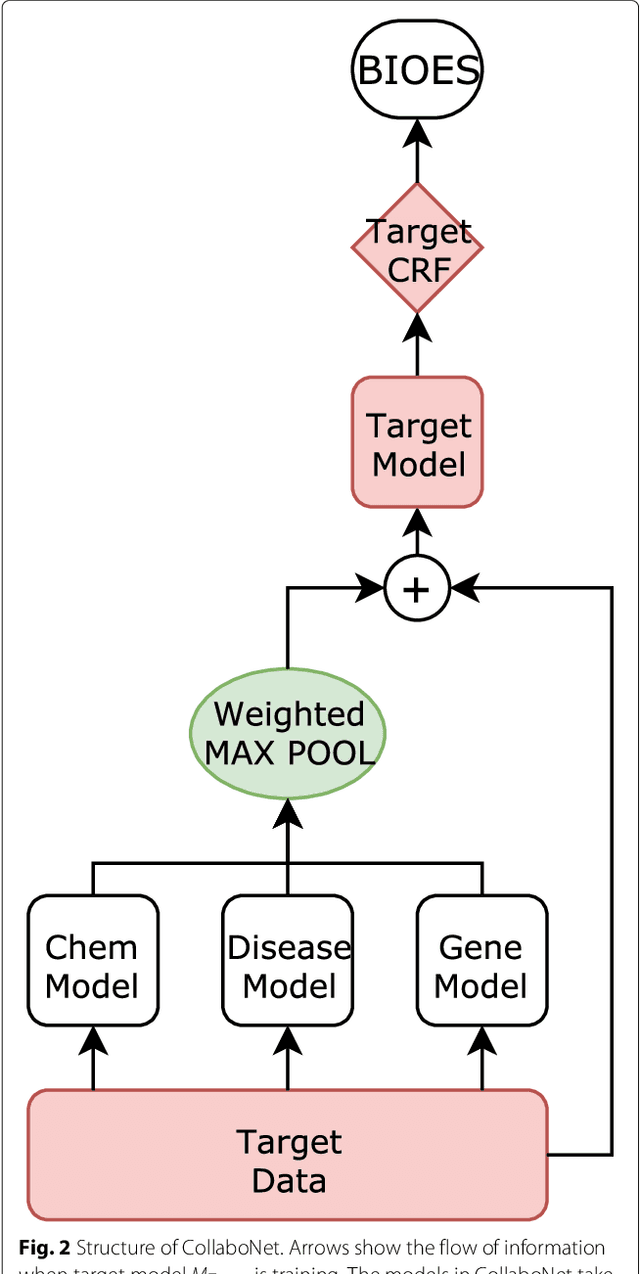
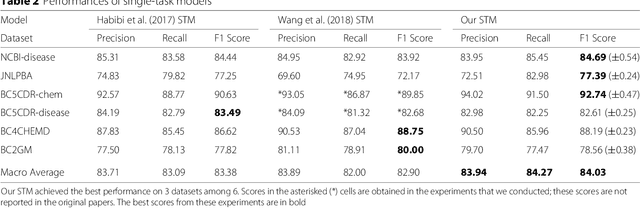
Abstract:Background: Finding biomedical named entities is one of the most essential tasks in biomedical text mining. Recently, deep learning-based approaches have been applied to biomedical named entity recognition (BioNER) and showed promising results. However, as deep learning approaches need an abundant amount of training data, a lack of data can hinder performance. BioNER datasets are scarce resources and each dataset covers only a small subset of entity types. Furthermore, many bio entities are polysemous, which is one of the major obstacles in named entity recognition. Results: To address the lack of data and the entity type misclassification problem, we propose CollaboNet which utilizes a combination of multiple NER models. In CollaboNet, models trained on a different dataset are connected to each other so that a target model obtains information from other collaborator models to reduce false positives. Every model is an expert on their target entity type and takes turns serving as a target and a collaborator model during training time. The experimental results show that CollaboNet can be used to greatly reduce the number of false positives and misclassified entities including polysemous words. CollaboNet achieved state-of-the-art performance in terms of precision, recall and F1 score. Conclusions: We demonstrated the benefits of combining multiple models for BioNER. Our model has successfully reduced the number of misclassified entities and improved the performance by leveraging multiple datasets annotated for different entity types. Given the state-of-the-art performance of our model, we believe that CollaboNet can improve the accuracy of downstream biomedical text mining applications such as bio-entity relation extraction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge