Wonjin Yoon
Using tournaments to calculate AUROC for zero-shot classification with LLMs
Feb 20, 2025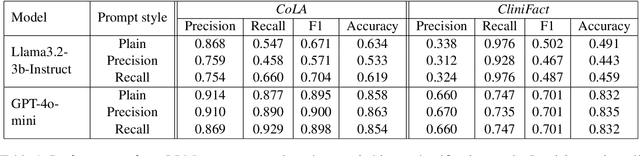
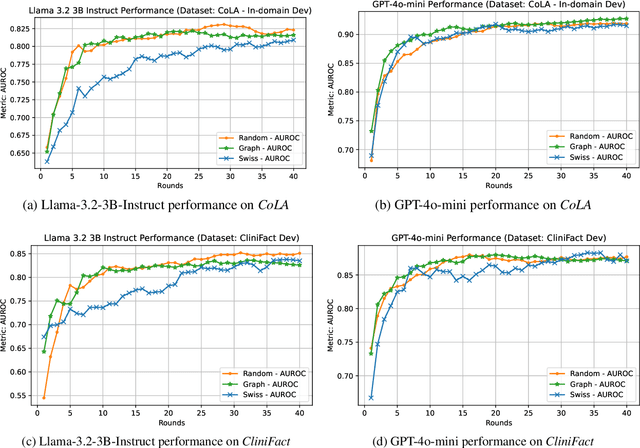
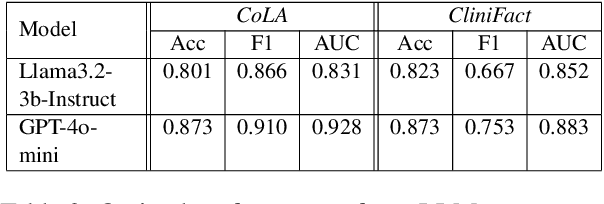
Abstract:Large language models perform surprisingly well on many zero-shot classification tasks, but are difficult to fairly compare to supervised classifiers due to the lack of a modifiable decision boundary. In this work, we propose and evaluate a method that converts binary classification tasks into pairwise comparison tasks, obtaining relative rankings from LLMs. Repeated pairwise comparisons can be used to score instances using the Elo rating system (used in chess and other competitions), inducing a confidence ordering over instances in a dataset. We evaluate scheduling algorithms for their ability to minimize comparisons, and show that our proposed algorithm leads to improved classification performance, while also providing more information than traditional zero-shot classification.
Biomedical NER for the Enterprise with Distillated BERN2 and the Kazu Framework
Dec 01, 2022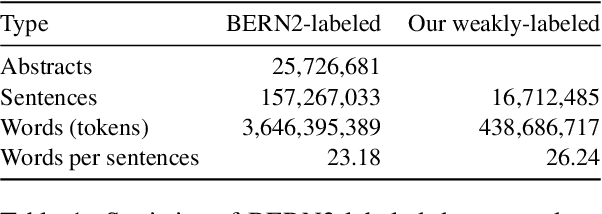
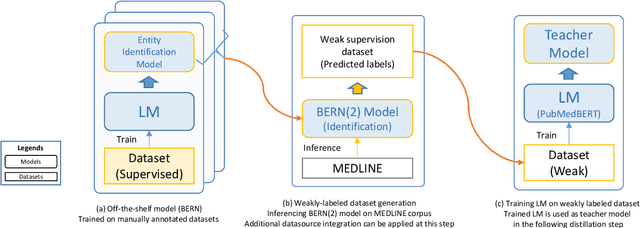
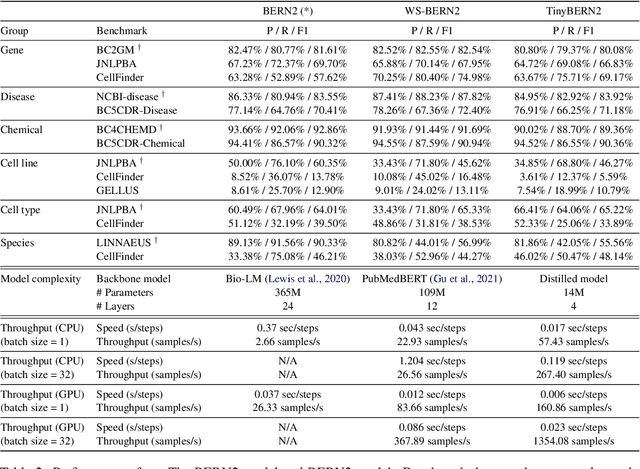
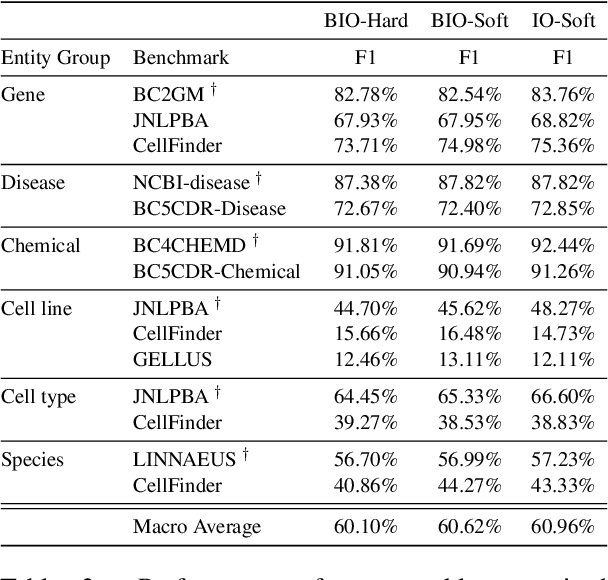
Abstract:In order to assist the drug discovery/development process, pharmaceutical companies often apply biomedical NER and linking techniques over internal and public corpora. Decades of study of the field of BioNLP has produced a plethora of algorithms, systems and datasets. However, our experience has been that no single open source system meets all the requirements of a modern pharmaceutical company. In this work, we describe these requirements according to our experience of the industry, and present Kazu, a highly extensible, scalable open source framework designed to support BioNLP for the pharmaceutical sector. Kazu is a built around a computationally efficient version of the BERN2 NER model (TinyBERN2), and subsequently wraps several other BioNLP technologies into one coherent system. KAZU framework is open-sourced: https://github.com/AstraZeneca/KAZU
Improving Tagging Consistency and Entity Coverage for Chemical Identification in Full-text Articles
Nov 20, 2021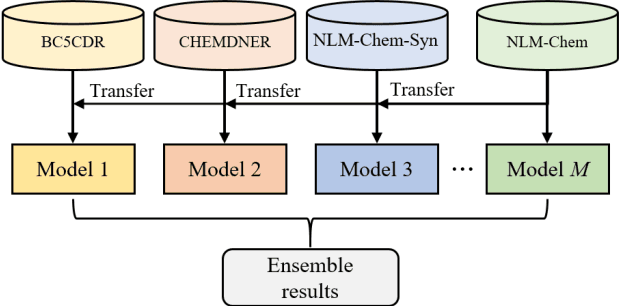
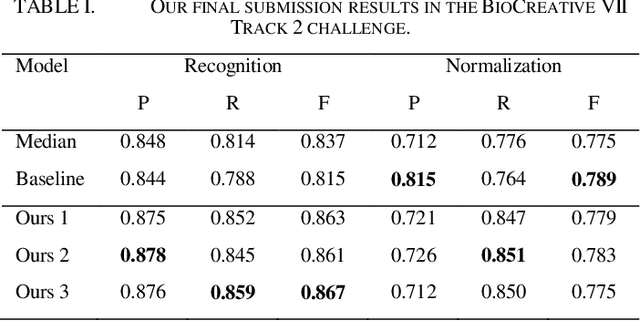
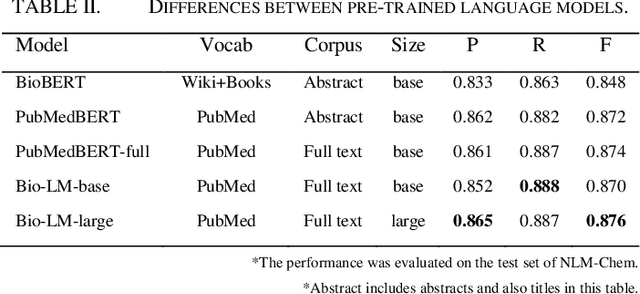
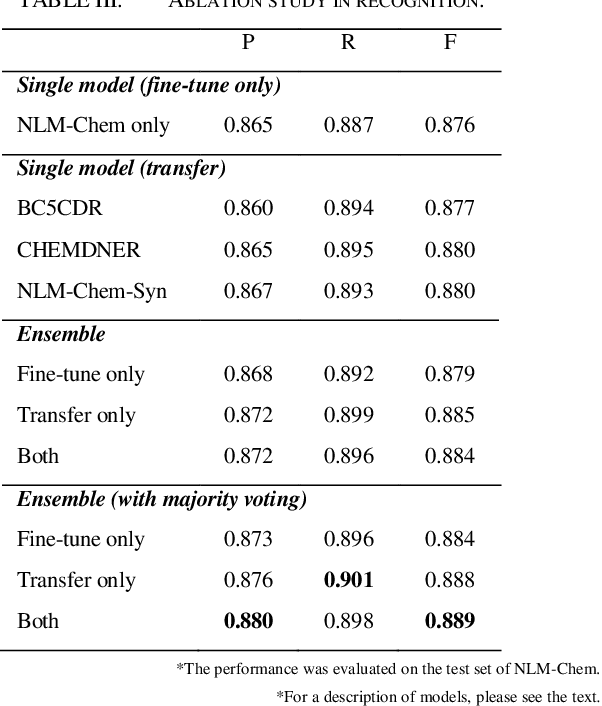
Abstract:This paper is a technical report on our system submitted to the chemical identification task of the BioCreative VII Track 2 challenge. The main feature of this challenge is that the data consists of full-text articles, while current datasets usually consist of only titles and abstracts. To effectively address the problem, we aim to improve tagging consistency and entity coverage using various methods such as majority voting within the same articles for named entity recognition (NER) and a hybrid approach that combines a dictionary and a neural model for normalization. In the experiments on the NLM-Chem dataset, we show that our methods improve models' performance, particularly in terms of recall. Finally, in the official evaluation of the challenge, our system was ranked 1st in NER by significantly outperforming the baseline model and more than 80 submissions from 16 teams.
Sequence Tagging for Biomedical Extractive Question Answering
Apr 15, 2021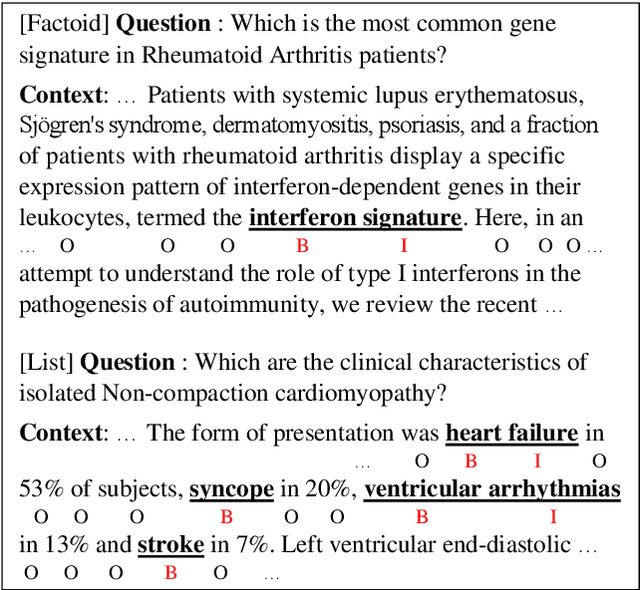
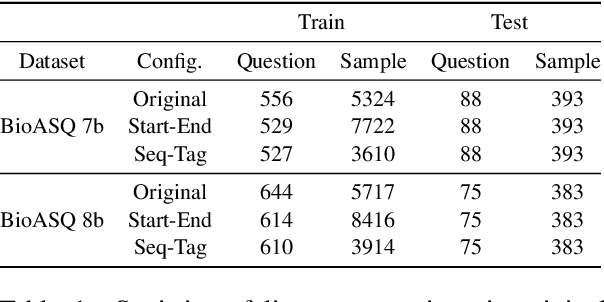
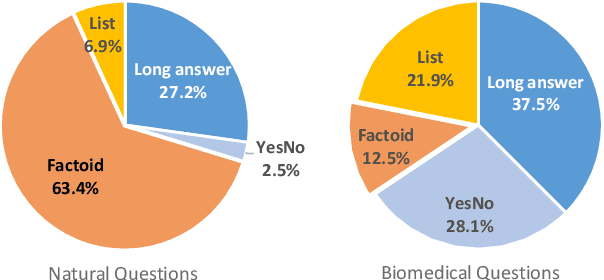
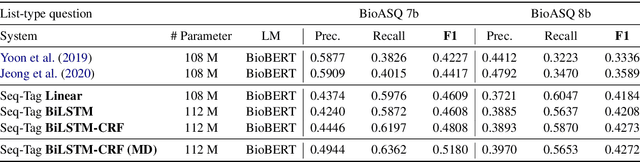
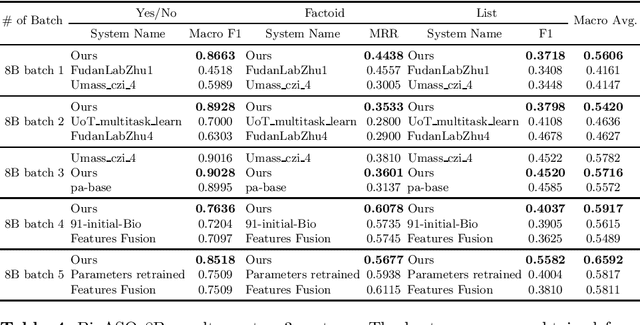
Abstract:Current studies in extractive question answering (EQA) have modeled single-span extraction setting, where a single answer span is a label to predict for a given question-passage pair. This setting is natural for general domain EQA as the majority of the questions in the general domain can be answered with a single span. Following general domain EQA models, current biomedical EQA (BioEQA) models utilize single-span extraction setting with post-processing steps. In this paper, we investigate the difference of the question distribution across the general and biomedical domains and discover biomedical questions are more likely to require list-type answers (multiple answers) than factoid-type answers (single answer). In real-world use cases, this emphasizes the need for Biomedical EQA models able to handle multiple question types. Based on this preliminary study, we propose a multi-span extraction setting, namely sequence tagging approach for BioEQA, which directly tackles questions with a variable number of phrases as their answer. Our approach can learn to decide the number of answers for a question from training data. Our experimental result on the BioASQ 7b and 8b list-type questions outperformed the best-performing existing models without requiring post-processing steps.
Transferability of Natural Language Inference to Biomedical Question Answering
Jul 01, 2020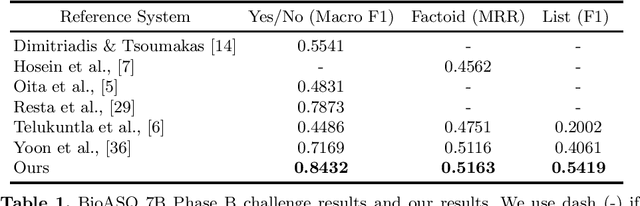
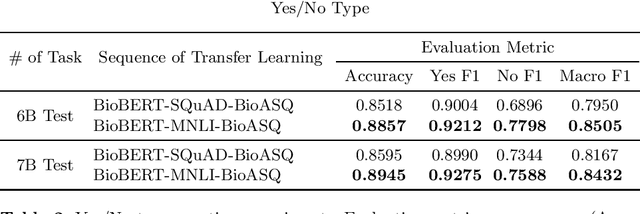
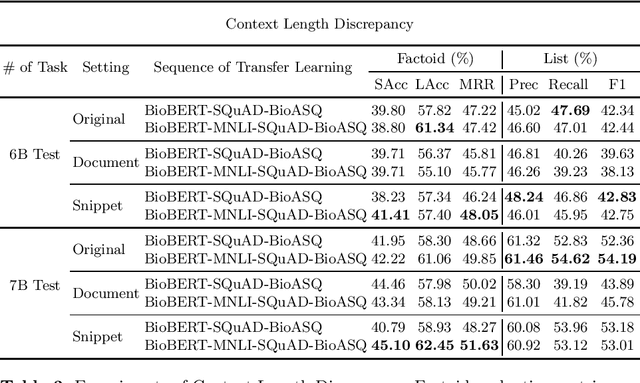
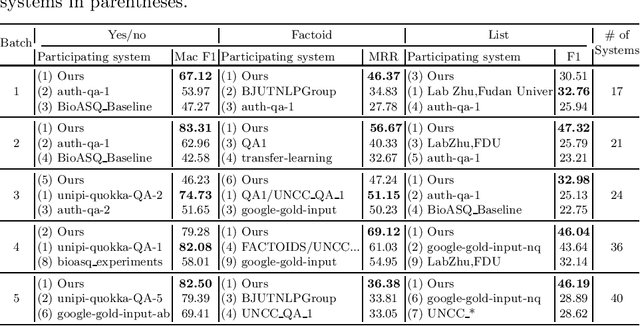
Abstract:Biomedical question answering (QA) is a challenging problem due to the scarcity of data and the requirement of domain expertise. Growing interests of using pre-trained language models with transfer learning address the issue to some extent. Recently, learning linguistic knowledge of entailment in sentence pairs enhances the performance in general domain QA by leveraging such transferability between the two tasks. In this paper, we focus on facilitating the transferability by unifying the experimental setup from natural language inference (NLI) to biomedical QA. We observe that transferring from entailment data shows effective performance on Yes/No (+5.59%), Factoid (+0.53%), List (+13.58%) type questions compared to previous challenge reports (BioASQ 7B Phase B). We also observe that our method generally performs well in the 8th BioASQ Challenge (Phase B). For sequential transfer learning, the order of how tasks are fine-tuned is important. In factoid- and list-type questions, we thoroughly analyze an intrinsic limitation of the extractive QA setting when these questions are converted to the same format of the Stanford Question Answering Dataset (SQuAD).
Answering Questions on COVID-19 in Real-Time
Jun 29, 2020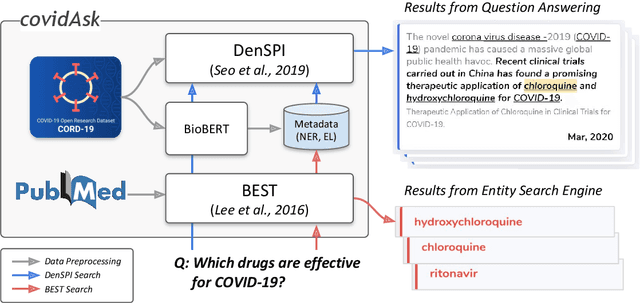
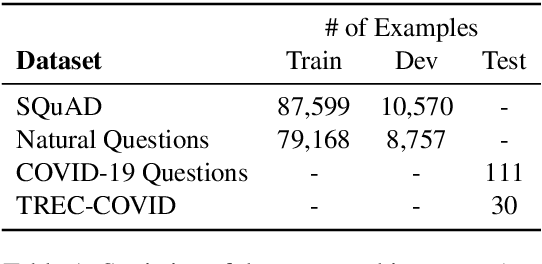
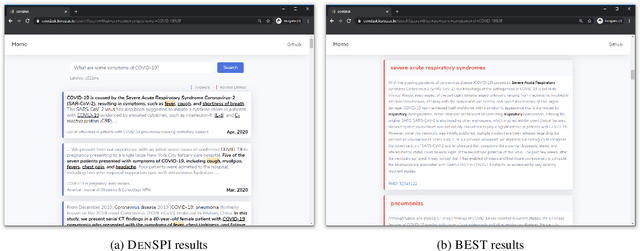

Abstract:The recent outbreak of the novel coronavirus is wreaking havoc on the world and researchers are struggling to effectively combat it. One reason why the fight is difficult is due to the lack of information and knowledge. In this work, we outline our effort to contribute to shrinking this knowledge vacuum by creating covidAsk, a question answering (QA) system that combines biomedical text mining and QA techniques to provide answers to questions in real-time. Our system leverages both supervised and unsupervised approaches to provide informative answers using DenSPI (Seo et al., 2019) and BEST (Lee et al., 2016). Evaluation of covidAsk is carried out by using a manually created dataset called COVID-19 Questions which is based on facts about COVID-19. We hope our system will be able to aid researchers in their search for knowledge and information not only for COVID-19 but for future pandemics as well.
Learning by Semantic Similarity Makes Abstractive Summarization Better
Feb 18, 2020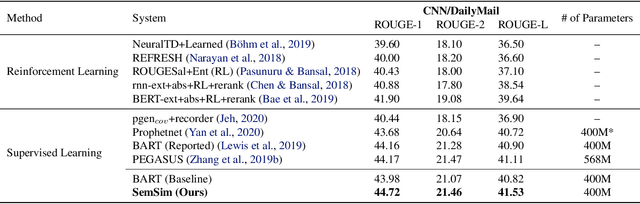
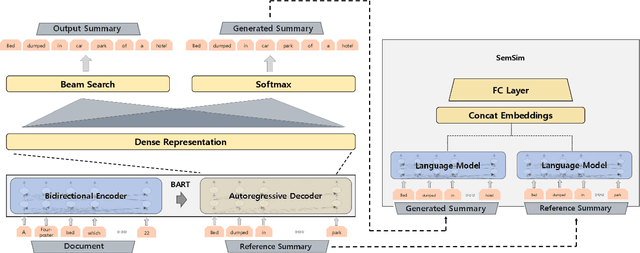
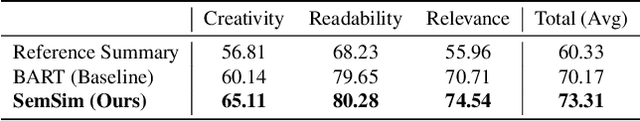

Abstract:One of the obstacles of abstractive summarization is the presence of various potentially correct predictions. Widely used objective functions for supervised learning, such as cross-entropy loss, cannot handle alternative answers effectively. Rather, they act as a training noise. In this paper, we propose Semantic Similarity strategy that can consider semantic meanings of generated summaries while training. Our training objective includes maximizing semantic similarity score which is calculated by an additional layer that estimates semantic similarity between generated summary and reference summary. By leveraging pre-trained language models, our model achieves a new state-of-the-art performance, ROUGE-L score of 41.5 on CNN/DM dataset. To support automatic evaluation, we also conducted human evaluation and received higher scores relative to both baseline and reference summaries.
Pre-trained Language Model for Biomedical Question Answering
Sep 18, 2019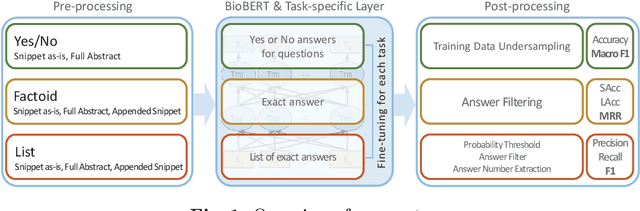
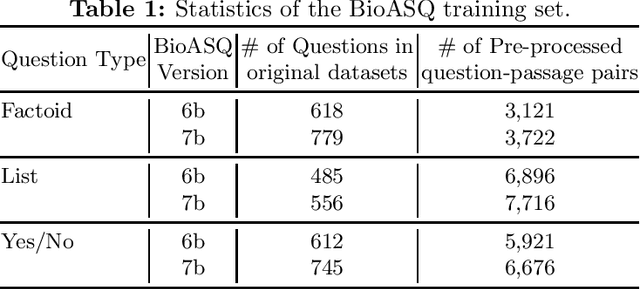
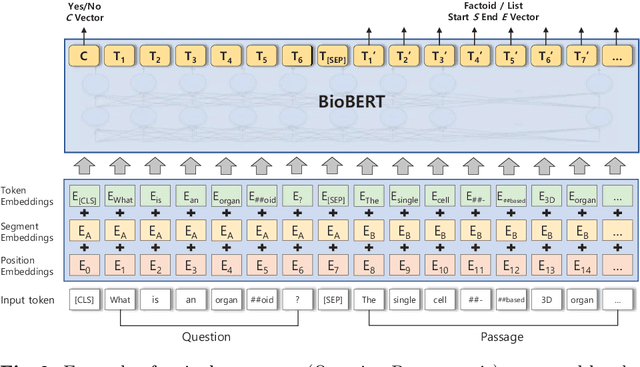
Abstract:The recent success of question answering systems is largely attributed to pre-trained language models. However, as language models are mostly pre-trained on general domain corpora such as Wikipedia, they often have difficulty in understanding biomedical questions. In this paper, we investigate the performance of BioBERT, a pre-trained biomedical language model, in answering biomedical questions including factoid, list, and yes/no type questions. BioBERT uses almost the same structure across various question types and achieved the best performance in the 7th BioASQ Challenge (Task 7b, Phase B). BioBERT pre-trained on SQuAD or SQuAD 2.0 easily outperformed previous state-of-the-art models. BioBERT obtains the best performance when it uses the appropriate pre-/post-processing strategies for questions, passages, and answers.
BioBERT: a pre-trained biomedical language representation model for biomedical text mining
Feb 03, 2019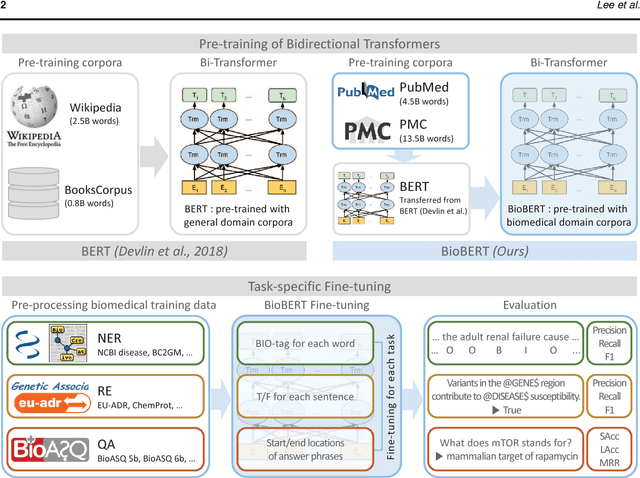
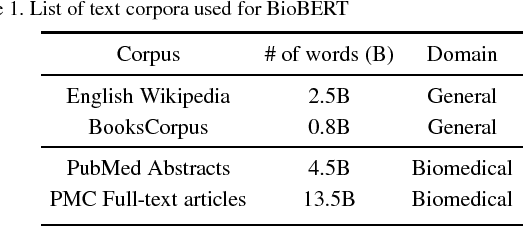

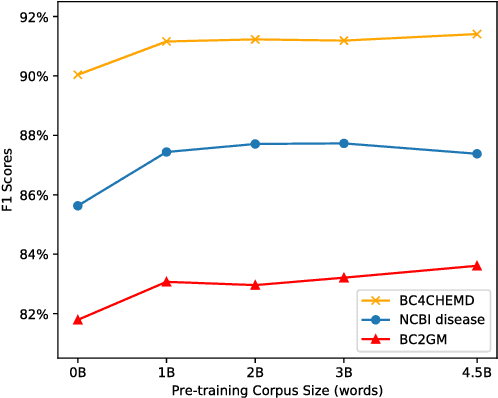
Abstract:Biomedical text mining is becoming increasingly important as the number of biomedical documents rapidly grows. With the progress in machine learning, extracting valuable information from biomedical literature has gained popularity among researchers, and deep learning has boosted the development of effective biomedical text mining models. However, as deep learning models require a large amount of training data, applying deep learning to biomedical text mining is often unsuccessful due to the lack of training data in biomedical fields. Recent researches on training contextualized language representation models on text corpora shed light on the possibility of leveraging a large number of unannotated biomedical text corpora. We introduce BioBERT (Bidirectional Encoder Representations from Transformers for Biomedical Text Mining), which is a domain specific language representation model pre-trained on large-scale biomedical corpora. Based on the BERT architecture, BioBERT effectively transfers the knowledge from a large amount of biomedical texts to biomedical text mining models with minimal task-specific architecture modifications. While BERT shows competitive performances with previous state-of-the-art models, BioBERT significantly outperforms them on the following three representative biomedical text mining tasks: biomedical named entity recognition (0.51% absolute improvement), biomedical relation extraction (3.49% absolute improvement), and biomedical question answering (9.61% absolute improvement). We make the pre-trained weights of BioBERT freely available at https://github.com/naver/biobert-pretrained, and the source code for fine-tuning BioBERT available at https://github.com/dmis-lab/biobert.
CollaboNet: collaboration of deep neural networks for biomedical named entity recognition
Sep 21, 2018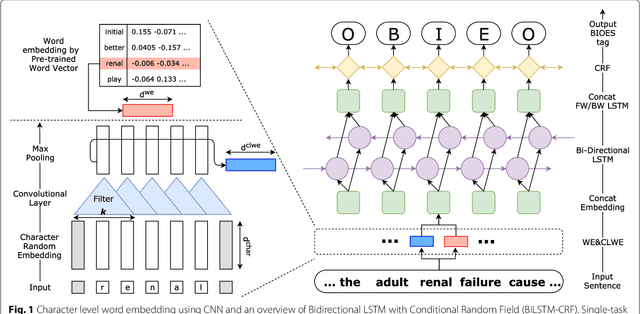
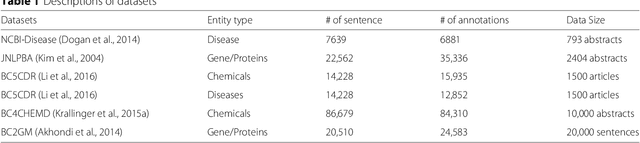
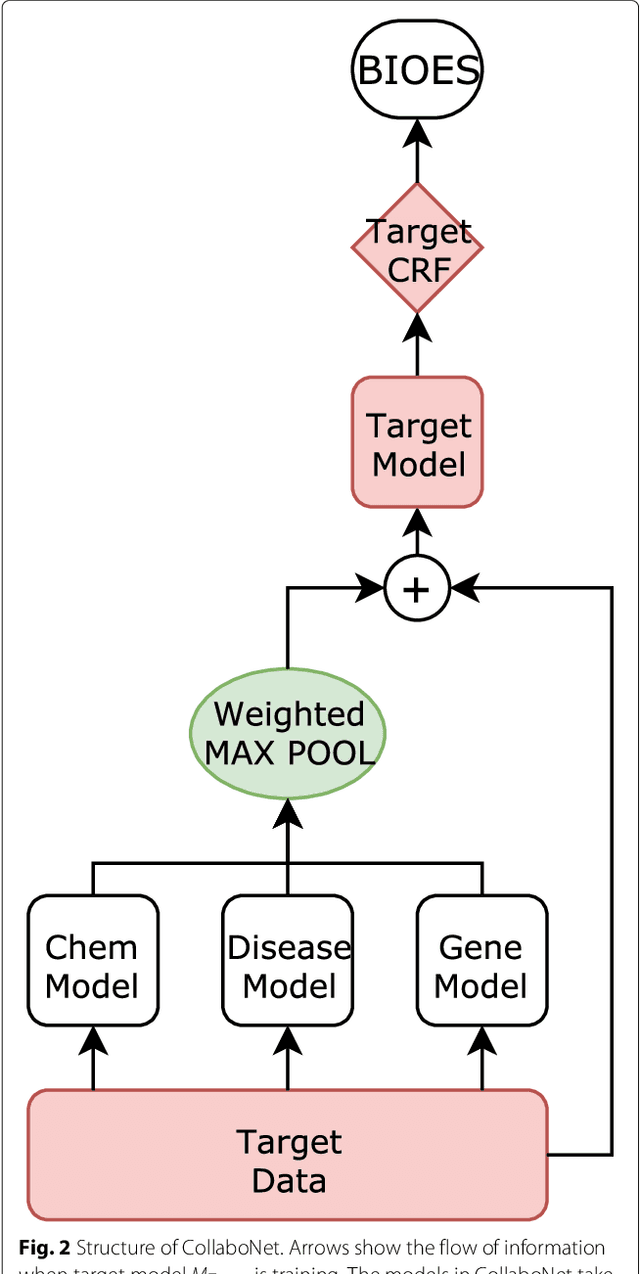
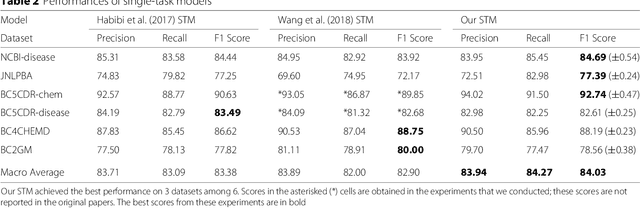
Abstract:Background: Finding biomedical named entities is one of the most essential tasks in biomedical text mining. Recently, deep learning-based approaches have been applied to biomedical named entity recognition (BioNER) and showed promising results. However, as deep learning approaches need an abundant amount of training data, a lack of data can hinder performance. BioNER datasets are scarce resources and each dataset covers only a small subset of entity types. Furthermore, many bio entities are polysemous, which is one of the major obstacles in named entity recognition. Results: To address the lack of data and the entity type misclassification problem, we propose CollaboNet which utilizes a combination of multiple NER models. In CollaboNet, models trained on a different dataset are connected to each other so that a target model obtains information from other collaborator models to reduce false positives. Every model is an expert on their target entity type and takes turns serving as a target and a collaborator model during training time. The experimental results show that CollaboNet can be used to greatly reduce the number of false positives and misclassified entities including polysemous words. CollaboNet achieved state-of-the-art performance in terms of precision, recall and F1 score. Conclusions: We demonstrated the benefits of combining multiple models for BioNER. Our model has successfully reduced the number of misclassified entities and improved the performance by leveraging multiple datasets annotated for different entity types. Given the state-of-the-art performance of our model, we believe that CollaboNet can improve the accuracy of downstream biomedical text mining applications such as bio-entity relation extraction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge