Improving Tagging Consistency and Entity Coverage for Chemical Identification in Full-text Articles
Paper and Code
Nov 20, 2021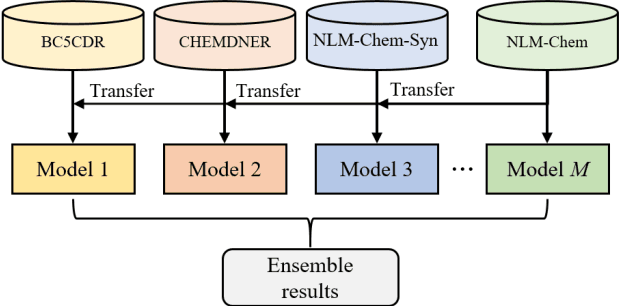
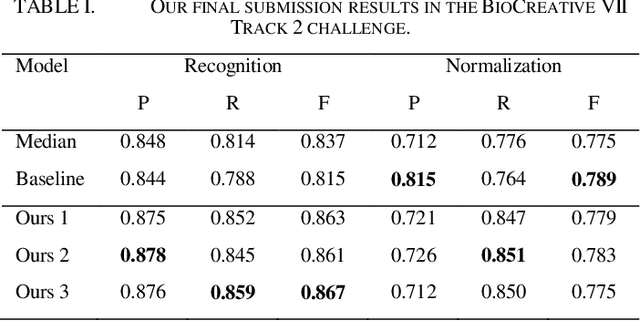
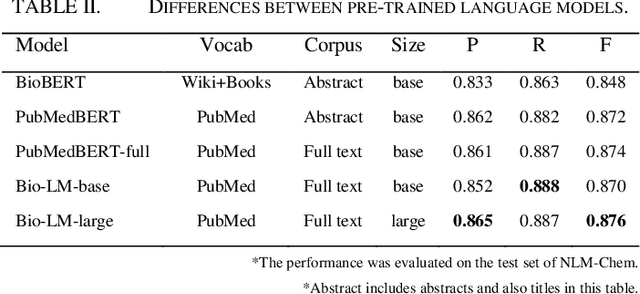
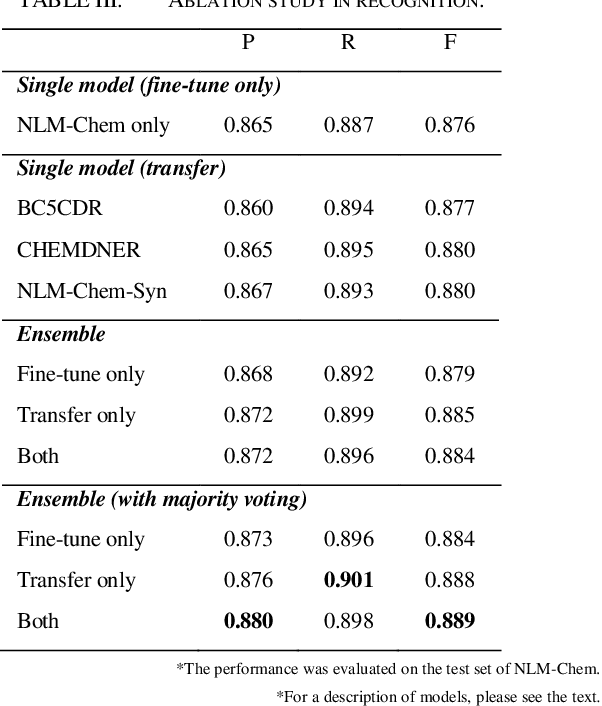
This paper is a technical report on our system submitted to the chemical identification task of the BioCreative VII Track 2 challenge. The main feature of this challenge is that the data consists of full-text articles, while current datasets usually consist of only titles and abstracts. To effectively address the problem, we aim to improve tagging consistency and entity coverage using various methods such as majority voting within the same articles for named entity recognition (NER) and a hybrid approach that combines a dictionary and a neural model for normalization. In the experiments on the NLM-Chem dataset, we show that our methods improve models' performance, particularly in terms of recall. Finally, in the official evaluation of the challenge, our system was ranked 1st in NER by significantly outperforming the baseline model and more than 80 submissions from 16 teams.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge