Vibha Anand
CTBench: A Comprehensive Benchmark for Evaluating Language Model Capabilities in Clinical Trial Design
Jun 25, 2024Abstract:CTBench is introduced as a benchmark to assess language models (LMs) in aiding clinical study design. Given study-specific metadata, CTBench evaluates AI models' ability to determine the baseline features of a clinical trial (CT), which include demographic and relevant features collected at the trial's start from all participants. These baseline features, typically presented in CT publications (often as Table 1), are crucial for characterizing study cohorts and validating results. Baseline features, including confounders and covariates, are also necessary for accurate treatment effect estimation in studies involving observational data. CTBench consists of two datasets: "CT-Repo," containing baseline features from 1,690 clinical trials sourced from clinicaltrials.gov, and "CT-Pub," a subset of 100 trials with more comprehensive baseline features gathered from relevant publications. Two LM-based evaluation methods are developed to compare the actual baseline feature lists against LM-generated responses. "ListMatch-LM" and "ListMatch-BERT" use GPT-4o and BERT scores (at various thresholds), respectively, for evaluation. To establish baseline results, advanced prompt engineering techniques using LLaMa3-70B-Instruct and GPT-4o in zero-shot and three-shot learning settings are applied to generate potential baseline features. The performance of GPT-4o as an evaluator is validated through human-in-the-loop evaluations on the CT-Pub dataset, where clinical experts confirm matches between actual and LM-generated features. The results highlight a promising direction with significant potential for improvement, positioning CTBench as a useful tool for advancing research on AI in CT design and potentially enhancing the efficacy and robustness of CTs.
Sparsity-based Feature Selection for Anomalous Subgroup Discovery
Jan 06, 2022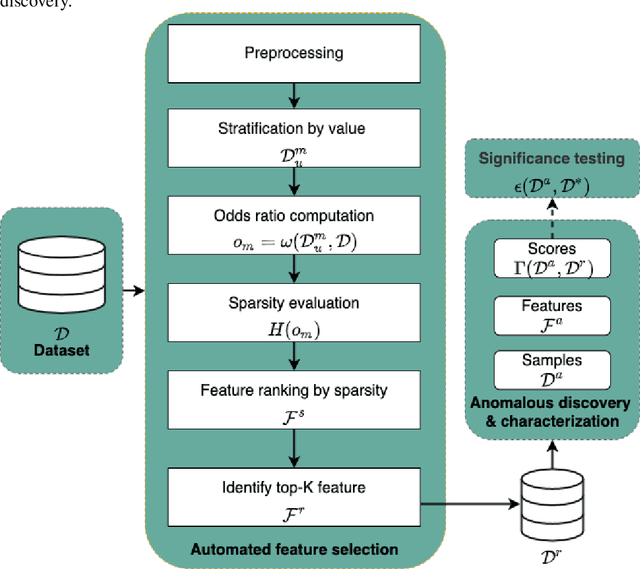
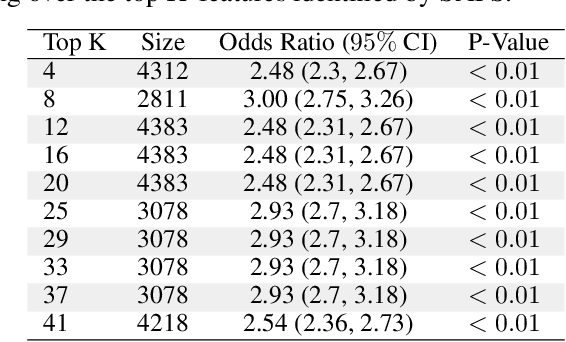
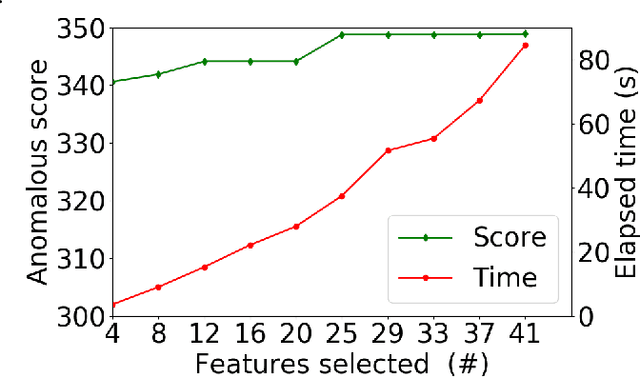
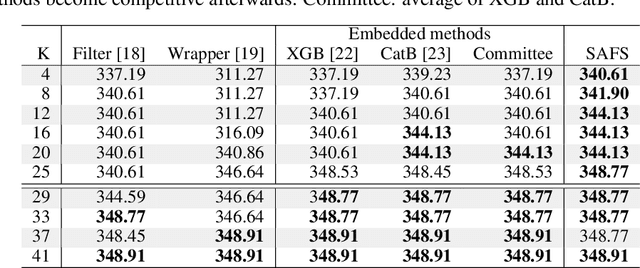
Abstract:Anomalous pattern detection aims to identify instances where deviation from normalcy is evident, and is widely applicable across domains. Multiple anomalous detection techniques have been proposed in the state of the art. However, there is a common lack of a principled and scalable feature selection method for efficient discovery. Existing feature selection techniques are often conducted by optimizing the performance of prediction outcomes rather than its systemic deviations from the expected. In this paper, we proposed a sparsity-based automated feature selection (SAFS) framework, which encodes systemic outcome deviations via the sparsity of feature-driven odds ratios. SAFS is a model-agnostic approach with usability across different discovery techniques. SAFS achieves more than $3\times$ reduction in computation time while maintaining detection performance when validated on publicly available critical care dataset. SAFS also results in a superior performance when compared against multiple baselines for feature selection.
Modeling Disease Progression Trajectories from Longitudinal Observational Data
Dec 09, 2020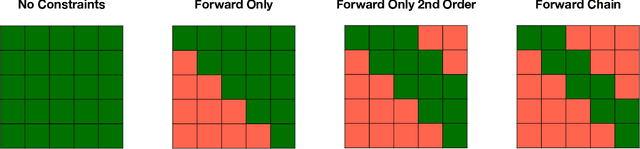

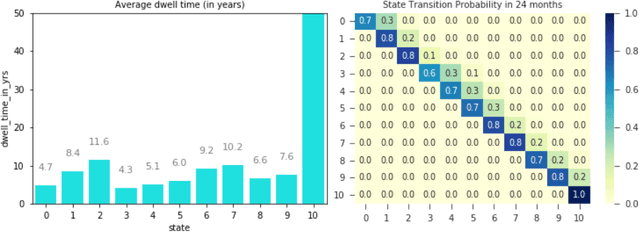
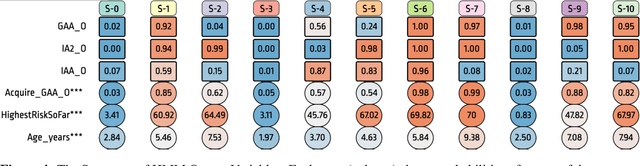
Abstract:Analyzing disease progression patterns can provide useful insights into the disease processes of many chronic conditions. These analyses may help inform recruitment for prevention trials or the development and personalization of treatments for those affected. We learn disease progression patterns using Hidden Markov Models (HMM) and distill them into distinct trajectories using visualization methods. We apply it to the domain of Type 1 Diabetes (T1D) using large longitudinal observational data from the T1DI study group. Our method discovers distinct disease progression trajectories that corroborate with recently published findings. In this paper, we describe the iterative process of developing the model. These methods may also be applied to other chronic conditions that evolve over time.
DPVis: Visual Exploration of Disease Progression Pathways
Apr 26, 2019Abstract:Clinical researchers use disease progression modeling algorithms to predict future patient status and characterize progression patterns. One approach for disease progression modeling is to describe patient status using a small number of states that represent distinctive distributions over a set of observed measures. Hidden Markov models (HMMs) and its variants are a class of models that both discover these states and make predictions concerning future states for new patients. HMMs can be trained using longitudinal observations of subjects from large-scale cohort studies, clinical trials, and electronic health records. Despite the advantages of using the algorithms for discovering interesting patterns, it still remains challenging for medical experts to interpret model outputs, complex modeling parameters, and clinically make sense of the patterns. To tackle this problem, we conducted a design study with physician scientists, statisticians, and visualization experts, with the goal to investigate disease progression pathways of certain chronic diseases, namely type 1 diabetes (T1D), Huntington's disease, Parkinson's disease, and chronic obstructive pulmonary disease (COPD). As a result, we introduce DPVis which seamlessly integrates model parameters and outcomes of HMMs into interpretable, and interactive visualizations. In this study, we demonstrate that DPVis is successful in evaluating disease progression models, visually summarizing disease states, interactively exploring disease progression patterns, and designing and comparing clinically relevant subgroup cohorts by introducing a case study on observation data from clinical studies of T1D.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge