Charles Wachira
Sparsity-based Feature Selection for Anomalous Subgroup Discovery
Jan 06, 2022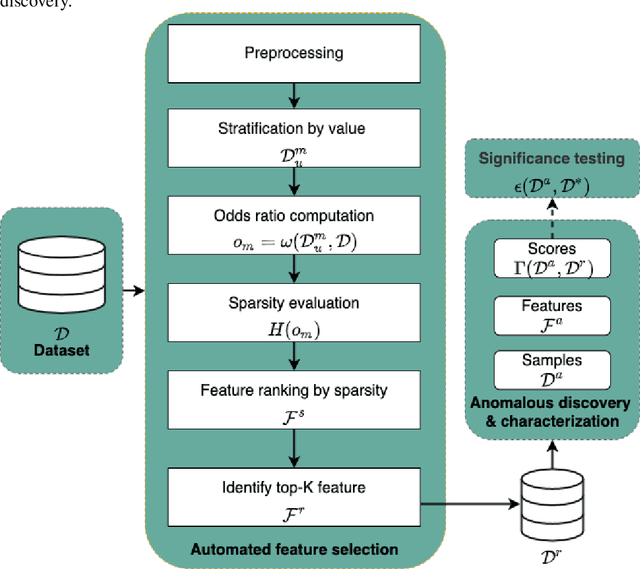
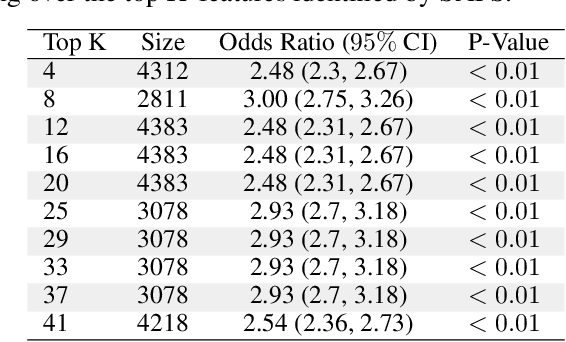
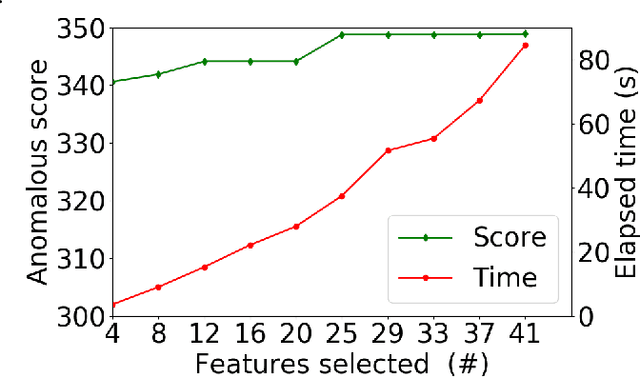
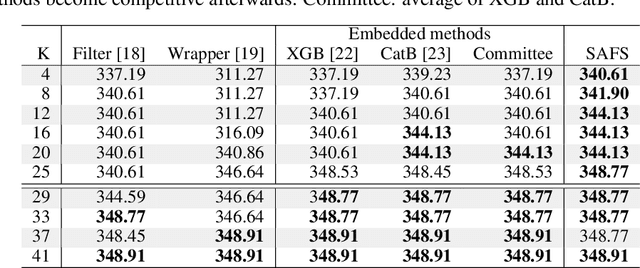
Abstract:Anomalous pattern detection aims to identify instances where deviation from normalcy is evident, and is widely applicable across domains. Multiple anomalous detection techniques have been proposed in the state of the art. However, there is a common lack of a principled and scalable feature selection method for efficient discovery. Existing feature selection techniques are often conducted by optimizing the performance of prediction outcomes rather than its systemic deviations from the expected. In this paper, we proposed a sparsity-based automated feature selection (SAFS) framework, which encodes systemic outcome deviations via the sparsity of feature-driven odds ratios. SAFS is a model-agnostic approach with usability across different discovery techniques. SAFS achieves more than $3\times$ reduction in computation time while maintaining detection performance when validated on publicly available critical care dataset. SAFS also results in a superior performance when compared against multiple baselines for feature selection.
Automated Supervised Feature Selection for Differentiated Patterns of Care
Nov 05, 2021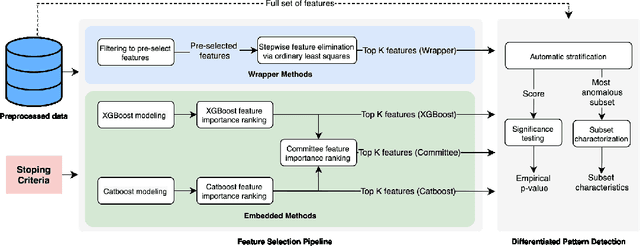
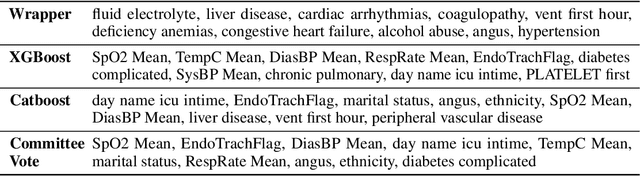
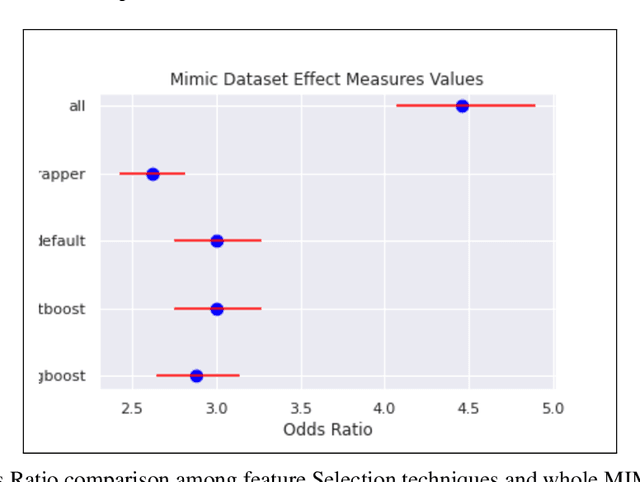
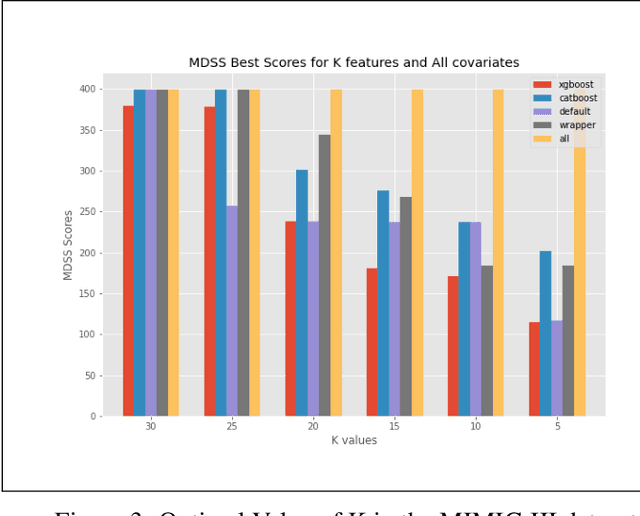
Abstract:An automated feature selection pipeline was developed using several state-of-the-art feature selection techniques to select optimal features for Differentiating Patterns of Care (DPOC). The pipeline included three types of feature selection techniques; Filters, Wrappers and Embedded methods to select the top K features. Five different datasets with binary dependent variables were used and their different top K optimal features selected. The selected features were tested in the existing multi-dimensional subset scanning (MDSS) where the most anomalous subpopulations, most anomalous subsets, propensity scores, and effect of measures were recorded to test their performance. This performance was compared with four similar metrics gained after using all covariates in the dataset in the MDSS pipeline. We found out that despite the different feature selection techniques used, the data distribution is key to note when determining the technique to use.
WNTRAC: Artificial Intelligence Assisted Tracking of Non-pharmaceutical Interventions Implemented Worldwide for COVID-19
Sep 16, 2020



Abstract:The Coronavirus disease 2019 (COVID-19) global pandemic has transformed almost every facet of human society throughout the world. Against an emerging, highly transmissible disease with no definitive treatment or vaccine, governments worldwide have implemented non-pharmaceutical intervention (NPI) to slow the spread of the virus. Examples of such interventions include community actions (e.g. school closures, restrictions on mass gatherings), individual actions (e.g. mask wearing, self-quarantine), and environmental actions (e.g. public facility cleaning). We present the Worldwide Non-pharmaceutical Interventions Tracker for COVID-19 (WNTRAC), a comprehensive dataset consisting of over 6,000 NPIs implemented worldwide since the start of the pandemic. WNTRAC covers NPIs implemented across 261 countries and territories, and classifies NPI measures into a taxonomy of sixteen NPI types. NPI measures are automatically extracted daily from Wikipedia articles using natural language processing techniques and manually validated to ensure accuracy and veracity. We hope that the dataset is valuable for policymakers, public health leaders, and researchers in modeling and analysis efforts for controlling the spread of COVID-19.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge